Comparison of Actinobacteria communities from human-impacted and pristine karst caves
- PMID: 35478281
- PMCID: PMC8988830
- DOI: 10.1002/mbo3.1276
Comparison of Actinobacteria communities from human-impacted and pristine karst caves
Abstract
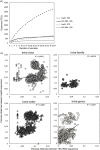
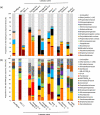
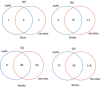
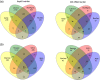
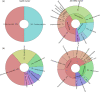
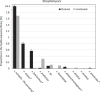
Actinobacteria are important cave inhabitants, but knowledge of how anthropization and anthropization-related visual marks affect this community on cave walls is lacking. We compared Actinobacteria communities among four French limestone caves (Mouflon, Reille, Rouffignac, and Lascaux) ranging from pristine to anthropized, and within Lascaux Cave between marked (wall visual marks) and unmarked areas in different rooms (Sas-1, Passage, Apse, and Diaclase). In addition to the 16S rRNA gene marker, 441 bp fragments of the hsp65 gene were used and an hsp65-related taxonomic database was constructed for the identification of Actinobacteria to the species level by Illumina-MiSeq analysis. The hsp65 marker revealed higher resolution for species and higher richness (99% operational taxonomic units cutoff) versus the 16S rRNA gene; however, more taxa were identified at higher taxonomic ranks. Actinobacteria communities varied between Mouflon and Reille caves (both pristine), and Rouffignac and Lascaux (both anthropized). Rouffignac displayed high diversity of Nocardia, suggesting human inputs, and Lascaux exhibited high Mycobacterium relative abundance, whereas Gaiellales were typical in pristine caves and the Diaclase (least affected area of Lascaux Cave). Within Lascaux, Pseudonocardiaceae dominated on unmarked walls and Streptomycetaceae (especially Streptomyces mirabilis) on marked walls, indicating a possible role in mark formation. A new taxonomic database was developed. Although not all Actinobacteria species were represented, the use of the hsp65 marker enabled species-level variations of the Actinobacteria community to be documented based on the extent of anthropogenic pressure. This approach proved effective when comparing different limestone caves or specific conditions within one cave.
Keywords: 16S rRNA gene sequencing; Actinobacteria; Paleolithic cave; cave anthropization; hsp65 sequencing; metabarcoding.
© 2022 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Conflict of interest statement
None declared.
Figures








References
-
- Aigle, A. , Colin, Y. , Bouchali, R. , Bourgeois, E. , Marti, R. , Ribun, S. , Marjolet, L. , Pozzi, A. C. M. , Misery, B. , Colinon, C. , Bernardin‐Souibgui, C. , Wiest, L. , Blaha, D. , Galia, W. , & Cournoyer, B. (2021). Spatio‐temporal variations in chemical pollutants found among urban deposits match changes in thiopurine S‐methyltransferase‐harboring bacteria tracked by the tpm metabarcoding approach. Science of the Total Environment, 767, 145425. 10.1016/j.scitotenv.2021 - DOI - PubMed
-
- Alonso, L. , Pommier, T. , Kaufmann, B. , Dubost, A. , Chapulliot, D. , Doré, J. , Douady, C. J. , & Moënne‐Loccoz, Y. (2019). Anthropization level of Lascaux Cave microbiome shown by regional‐scale comparisons of pristine and anthropized caves. Molecular Ecology, 28(14), 3383–3394. 10.1111/mec.15144 - DOI - PubMed
-
- Barraclough, T. G. , Balbi, K. J. , & Ellis, R. J. (2012). Evolving concepts of bacterial species. Evolution Biology, 39, 148–157. 10.1007/s11692-012-9181-8 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

