Recombinant Limosilactobacillus (Lactobacillus) delivering nanobodies against Clostridium perfringens NetB and alpha toxin confers potential protection from necrotic enteritis
- PMID: 35478283
- PMCID: PMC8924699
- DOI: 10.1002/mbo3.1270
Recombinant Limosilactobacillus (Lactobacillus) delivering nanobodies against Clostridium perfringens NetB and alpha toxin confers potential protection from necrotic enteritis
Abstract
Necrotic enteritis (NE), caused by Clostridium perfringens, is an intestinal disease with devastating economic losses to the poultry industry. NE is a complex disease and predisposing factors that compromise gut integrity are required to facilitate C. perfringens proliferation and toxin production. NE is also characterized by drastic shifts in gut microbiota; C. perfringens is negatively correlated with Lactobacilli. Vaccines are only partially effective against NE and antibiotics suffer from the concern of resistance development. These strategies address only some aspects of NE pathogenesis. Thus, there is an urgent need for alternative strategies that address multiple aspects of NE biology. Here, we developed Limosilactobacillus (Lactobacillus) reuteri vectors for in situ delivery of nanobodies against NetB and α toxin, two key toxins associated with NE pathophysiology. We generated nanobodies and showed that these nanobodies neutralize NetB and α toxin. We selected L. reuteri vector strains with intrinsic benefits and demonstrated that these strains inhibit C. perfringens and secrete over 130 metabolites, some of which play a key role in maintaining gut health. Recombinant L. reuteri strains efficiently secreted nanobodies and these nanobodies neutralized NetB. The recombinant strains were genetically and phenotypically stable over 480 generations and showed persistent colonization in chickens. A two-dose in ovo and drinking water administration of recombinant L. reuteri strains protected chickens from NE-associated mortality. These results provide proof-of-concept data for using L. reuteri as a live vector for delivery of nanobodies with broad applicability to other targets and highlight the potential synergistic effects of vector strains and nanobodies for addressing complex diseases such as NE.
Keywords: Limosilactobacillus; nanobodies; necrotic enteritis; poultry.
© 2022 The Authors. MicrobiologyOpen published by John Wiley & Sons Ltd.
Conflict of interest statement
Dharanesh Gangaiah, Valerie Ryan, Shrinivasrao P. Mane, Enid T. McKinley, Nandakumar D. Reddy, and Arvind Kumar are employees of Elanco Animal Health Inc. Nallakannu Lakshmanan was an employee of Elanco Animal Health Inc. at the time the work was done. Elanco Animal Health Inc. is a company that develops, manufactures, and sells veterinary pharmaceuticals and nutritionals. Daphne van Hoese and Edward Dolk are current employees of QVQ Holding BV, which discovers and develops nanobodies for different target diseases.
Figures




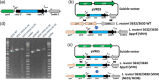


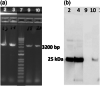
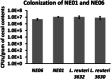
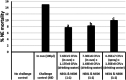




References
-
- Abildgaard, L. , Engberg, R. M. , Pedersen, K. , Schramm, A. , & Hojberg, O. (2009). Sequence variation in the alpha‐toxin encoding plc gene of Clostridium perfringens strains isolated from diseased and healthy chickens. Veterinary Microbiology, 136, 293–299. - PubMed
-
- Alimolaei, M. , Golchin, M. , & Ezatkhah, M. (2017). Orally administered recombinant Lactobacillus casei vector vaccine expressing beta‐toxoid of Clostridium perfringens that induced protective immunity responses. Research in Veterinary Science, 115, 332–339. - PubMed
-
- Allain, T. , Mansour, N. M. , Bahr, M. M. , Martin, R. , Florent, I. , Langella, P. , & Bermudez‐Humaran, L. G. (2016). A new lactobacilli in vivo expression system for the production and delivery of heterologous proteins at mucosal surfaces. FEMS Microbiology Letters, 363(13), fnw117. - PubMed
-
- Al‐Sheikhly, F. , & Truscott, R. B. (1977a). The pathology of necrotic enteritis of chickens following infusion of broth cultures of Clostridium perfringens into the duodenum. Avian Diseases, 21, 230–240. - PubMed
-
- Al‐Sheikhly, F. , & Truscott, R. B. (1977b). The pathology of necrotic enteritis of chickens following infusion of crude toxins of Clostridium perfringens into the duodenum. Avian Diseases, 21, 241–255. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

