Identification of Hub Genes in Idiopathic Pulmonary Fibrosis and NSCLC Progression:Evidence From Bioinformatics Analysis
- PMID: 35480306
- PMCID: PMC9038140
- DOI: 10.3389/fgene.2022.855789
Identification of Hub Genes in Idiopathic Pulmonary Fibrosis and NSCLC Progression:Evidence From Bioinformatics Analysis
Abstract
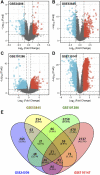
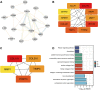
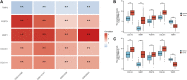
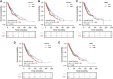
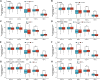
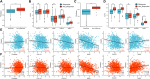
Background: Lung cancer is the most common comorbidity of idiopathic pulmonary fibrosis. Thus there is an urgent need for the research of IPF and carcinogenesis Objective: The objective of this study was to explore hub genes which are common in pulmonary fibrosis and lung cancer progression through bioinformatic analysis. Methods: All the analysis was performed in R software. Differentially expressed genes (DEGs) were explored by comparing gene expression profiles between IPF tissues and healthy lung tissues from GSE24206, GSE53845, GSE101286 and GSE110147 datasets. Venn Diagram analysis was used to identify the overlapping genes, while GO and KEGG pathway enrichment analysis were used to explore the biological functions of the DEGs using clusterprofiler package. Hub genes were identified by analyzing protein-protein interaction networks using Cytoscape software. Nomogram was constructed using the rms package. Tumor immune dysfunction and exclusion (TIDE) and Genomics of Drug Sensitivity in Cancer (GDSC) analysis was used to quantify the immunotherapy and chemotherapy sensitivity of non-small cell lung cancer (NSCLC) patients. Results: COL1A1, COL3A1, MMP1, POSTN1 and TIMP3 were identified as the top five hub genes. The five hub genes were used to construct a diagnostic nomogram that was validated in another IPF dataset. Since the hub genes were also associated with lung cancer progression, we found that the nomogram also had diagnostic value in NSCLC patients. These five genes achieved a statistically difference of overall survival in NSCLC patients (p < 0.05). The expression of the five hub genes was mostly enriched in fibroblasts. Fibroblasts and the hub genes also showed significant ability to predict the susceptibility of NSCLC patients to chemotherapy and immunotherapy. Conclusion: We identified five hub genes as potential biomarkers of IPF and NSCLC progression. This finding may give insight into the underlying molecular mechanisms of IPF and lung cancer progression and provides potential targets for developing new therapeutic agents for IPF patients.
Keywords: bioinformatics analysis; hub genes; idiopathic pulmonary fibrosis; lung cancer progression; pathways.
Copyright © 2022 Yao, Li and Gao.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Prediction and analysis of genetic effect in idiopathic pulmonary fibrosis and gastroesophageal reflux disease.IET Syst Biol. 2023 Dec;17(6):352-365. doi: 10.1049/syb2.12081. Epub 2023 Oct 31. IET Syst Biol. 2023. PMID: 37907428 Free PMC article.
-
Identification of Hub Genes in Idiopathic Pulmonary Fibrosis and Their Association with Lung Cancer by Bioinformatics Analysis.Adv Respir Med. 2023 Oct 12;91(5):407-431. doi: 10.3390/arm91050032. Adv Respir Med. 2023. PMID: 37887075 Free PMC article.
-
Bioinformatic analysis of differentially expressed genes and pathways in idiopathic pulmonary fibrosis.Ann Transl Med. 2021 Sep;9(18):1459. doi: 10.21037/atm-21-4224. Ann Transl Med. 2021. PMID: 34734011 Free PMC article.
-
Identification of oxidative stress-related diagnostic markers and immune infiltration features for idiopathic pulmonary fibrosis by bibliometrics and bioinformatics.Front Med (Lausanne). 2024 Aug 6;11:1356825. doi: 10.3389/fmed.2024.1356825. eCollection 2024. Front Med (Lausanne). 2024. PMID: 39165378 Free PMC article. Review.
-
Identification of the effects of COVID-19 on patients with pulmonary fibrosis and lung cancer: a bioinformatics analysis and literature review.Sci Rep. 2022 Sep 26;12(1):16040. doi: 10.1038/s41598-022-20040-x. Sci Rep. 2022. PMID: 36163484 Free PMC article. Review.
Cited by
-
Machine learning-based prediction of candidate gene biomarkers correlated with immune infiltration in patients with idiopathic pulmonary fibrosis.Front Med (Lausanne). 2023 Feb 13;10:1001813. doi: 10.3389/fmed.2023.1001813. eCollection 2023. Front Med (Lausanne). 2023. PMID: 36860337 Free PMC article.
-
Cellular therapies for idiopathic pulmonary fibrosis: current progress and future prospects.Am J Stem Cells. 2024 Aug 25;13(4):191-211. doi: 10.62347/DAKS5508. eCollection 2024. Am J Stem Cells. 2024. PMID: 39308764 Free PMC article. Review.
-
Lung transplantation in a woman with paraquat poisoning that led to pulmonary fibrosis-Widely reported by the media: A case report.Medicine (Baltimore). 2022 Dec 9;101(49):e32263. doi: 10.1097/MD.0000000000032263. Medicine (Baltimore). 2022. PMID: 36626514 Free PMC article.
-
Development of a Novel Biomarker for the Progression of Idiopathic Pulmonary Fibrosis.Int J Mol Sci. 2024 Jan 2;25(1):599. doi: 10.3390/ijms25010599. Int J Mol Sci. 2024. PMID: 38203769 Free PMC article.
-
Identification of Hub Genes and Prediction of Targeted Drugs for Rheumatoid Arthritis and Idiopathic Pulmonary Fibrosis.Biochem Genet. 2024 Dec;62(6):5157-5178. doi: 10.1007/s10528-023-10650-z. Epub 2024 Feb 9. Biochem Genet. 2024. PMID: 38334875
References
-
- Åhrman E., Hallgren O., Malmström L., Hedström U., Malmström A., Bjermer L., et al. (2018). Quantitative Proteomic Characterization of the Lung Extracellular Matrix in Chronic Obstructive Pulmonary Disease and Idiopathic Pulmonary Fibrosis. J. Proteomics 189, 23–33. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

