Structural Characterization of the Full-Length Anti-CD20 Antibody Rituximab
- PMID: 35480889
- PMCID: PMC9037831
- DOI: 10.3389/fmolb.2022.823174
Structural Characterization of the Full-Length Anti-CD20 Antibody Rituximab
Abstract
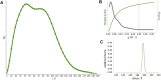
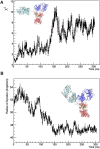
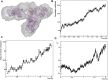
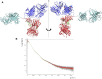
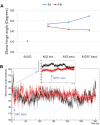
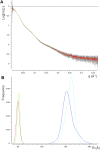
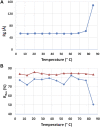
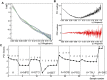
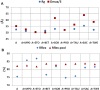
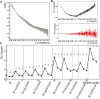
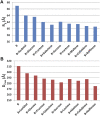
Rituximab, a murine-human chimera, is the first monoclonal antibody (mAb) developed as a therapeutic agent to target CD20 protein. Its Fab domain and its interaction with CD20 have been extensively studied and high-resolution atomic models obtained by X-ray diffraction or cryo-electron microscopy are available. However, the structure of the full-length antibody is still missing as the inherent protein flexibility hampers the formation of well-diffracting crystals and the reconstruction of 3D microscope images. The global structure of rituximab from its dilute solution is here elucidated by small-angle X-ray scattering (SAXS). The limited data resolution achievable by this technique has been compensated by intensive computational modelling that led to develop a new and effective procedure to characterize the average mAb conformation as well as that of the single domains. SAXS data indicated that rituximab adopts an asymmetric average conformation in solution, with a radius of gyration and a maximum linear dimension of 52 Å and 197 Å, respectively. The asymmetry is mainly due to an uneven arrangement of the two Fab units with respect to the central stem (the Fc domain) and reflects in a different conformation of the individual units. As a result, the Fab elbow angle, which is a crucial determinant for antigen recognition and binding, was found to be larger (169°) in the more distant Fab unit than that in the less distant one (143°). The whole flexibility of the antibody has been found to strongly depend on the relative inter-domain orientations, with one of the Fab arms playing a major role. The average structure and the amount of flexibility has been studied in the presence of different buffers and additives, and monitored at increasing temperature, up to the complete unfolding of the antibody. Overall, the structural characterization of rituximab can help in designing next-generation anti-CD20 antibodies and finding more efficient routes for rituximab production at industrial level.
Keywords: anti-CD20 mab; full-length antibody; molecular dynamics flexible fitting; small-angle X-ray scattering; structural comparison.
Copyright © 2022 Belviso, Mangiatordi, Alberga, Mangini, Carrozzini and Caliandro.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Adelman S. A., Doll J. D. (1976). Generalized Langevin Equation Approach for Atom/Solid-Surface Scattering: General Formulation for Classical Scattering off Harmonic Solids. J. Chem. Phys. 64, 2375–2388. 10.1063/1.432526 - DOI
-
- Alberga D., Trisciuzzi D., Lattanzi G., Bennett J. L., Verkman A. S., Mangiatordi G. F., et al. (2017). Comparative Molecular Dynamics Study of Neuromyelitis Optica-Immunoglobulin G Binding to Aquaporin-4 Extracellular Domains. Biochim. Biophys. Acta 1859, 1326–1334. 10.1016/j.bbamem.2017.05.001 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials

