Structural insights into DNA binding domain of vancomycin-resistance-associated response regulator in complex with its promoter DNA from Staphylococcus aureus
- PMID: 35481641
- PMCID: PMC8994486
- DOI: 10.1002/pro.4286
Structural insights into DNA binding domain of vancomycin-resistance-associated response regulator in complex with its promoter DNA from Staphylococcus aureus
Abstract
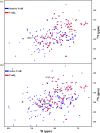
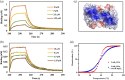
In Staphylococcus aureus, vancomycin-resistance-associated response regulator (VraR) is a part of the VraSR two-component system, which is responsible for activating a cell wall-stress stimulon in response to an antibiotic that inhibits cell wall formation. Two VraR-binding sites have been identified: R1 and R2 in the vraSR operon control region. However, the binding of VraR to a promoter DNA enhancing downstream gene expression remains unclear. VraR contains a conserved N-terminal receiver domain (VraRN ) connected to a C-terminal DNA binding domain (VraRC ) with a flexible linker. Here, we present the crystal structure of VraRC alone and in complex with R1-DNA in 1.87- and 2.0-Å resolution, respectively. VraRC consisting of four α-helices forms a dimer when interacting with R1-DNA. In the VraRC -DNA complex structure, Mg2+ ion is bound to Asp194. Biolayer interferometry experiments revealed that the addition of Mg2+ to VraRC enhanced its DNA binding affinity by eightfold. In addition, interpretation of NMR titrations between VraRC with R1- and R2-DNA revealed the essential residues that might play a crucial role in interacting with DNA of the vraSR operon. The structural information could help in designing and screening potential therapeutics/inhibitors to deal with antibiotic-resistant S. aureus via targeting VraR.
Keywords: DNA binding domain; NMR; Staphylococcus aureus; response regulator; two-component system; vancomycin resistance; x-ray crystal structure.
© 2022 The Protein Society.
Conflict of interest statement
The authors declare that there is no conflict of interest regarding the publication of this paper.
Figures


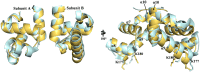



References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

