A Hybrid Adherent/Suspension Cell-Based Selection Strategy for Discovery of Antibodies Targeting Membrane Proteins
- PMID: 35482192
- PMCID: PMC9667817
- DOI: 10.1007/978-1-0716-2285-8_11
A Hybrid Adherent/Suspension Cell-Based Selection Strategy for Discovery of Antibodies Targeting Membrane Proteins
Abstract
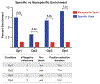
Membrane proteins are favored drug targets and antibody therapeutics represent the fastest-growing category of pharmaceuticals. However, there remains a need for rapid and effective approaches for the discovery of antibodies that recognize membrane proteins to develop a robust clinical pipeline for targeted therapeutics. The challenges associated with recombinant expression of membrane proteins make whole cell screening techniques desirable, as these strategies allow presentation of the target membrane proteins in their native conformations. Here, we describe a workflow that employs both adherent cell-based and suspension cell-based whole cell panning methodologies to enrich for specific binders within a yeast-displayed antibody library. The first round of selection consists of an adherent cell-based approach, wherein a diverse library is panned over target-expressing mammalian cell monolayers in order to debulk the naïve library. Subsequent rounds involve the use of suspension cell-based approaches, facilitated with magnetic-activated cell sorting (MACS) or fluorescence-activated cell sorting (FACS), to achieve further library enrichment. Finally, we describe a high-throughput approach to screen target binding and specificity of individual clones isolated from selection campaigns.
Keywords: Antibody discovery; Biopanning; Cell panning; Directed evolution; Membrane proteins; Molecular engineering; Yeast surface display.
© 2022. The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature.
Figures




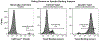
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

