Multiomic analysis of microRNA-mediated regulation reveals a proliferative axis involving miR-10b in fibrolamellar carcinoma
- PMID: 35482409
- PMCID: PMC9220943
- DOI: 10.1172/jci.insight.154743
Multiomic analysis of microRNA-mediated regulation reveals a proliferative axis involving miR-10b in fibrolamellar carcinoma
Abstract
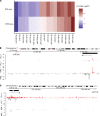
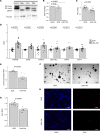
Fibrolamellar carcinoma (FLC) is an aggressive liver cancer primarily afflicting adolescents and young adults. Most patients with FLC harbor a heterozygous deletion on chromosome 19 that leads to the oncogenic gene fusion, DNAJB1-PRKACA. There are currently no effective therapeutics for FLC. To address that, it is critical to gain deeper mechanistic insight into FLC pathogenesis. We assembled a large sample set of FLC and nonmalignant liver tissue (n = 52) and performed integrative multiomic analysis. Specifically, we carried out small RNA sequencing to define altered microRNA expression patterns in tumor samples and then coupled this analysis with RNA sequencing and chromatin run-on sequencing data to identify candidate master microRNA regulators of gene expression in FLC. We also evaluated the relationship between DNAJB1-PRKACA and microRNAs of interest in several human and mouse cell models. Finally, we performed loss-of-function experiments for a specific microRNA in cells established from a patient-derived xenograft (PDX) model. We identified miR-10b-5p as the top candidate pro-proliferative microRNA in FLC. In multiple human cell models, overexpression of DNAJB1-PRKACA led to significant upregulation of miR-10b-5p. Inhibition of miR-10b in PDX-derived cells increased the expression of several potentially novel target genes, concomitant with a significant reduction in metabolic activity, proliferation, and anchorage-independent growth. This study highlights a potentially novel proliferative axis in FLC and provides a rich resource for further investigation of FLC etiology.
Keywords: Liver cancer; Molecular genetics; Oncogenes; Oncology.
Conflict of interest statement
Figures






References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

