Simulation and annotation of global acronyms
- PMID: 35482480
- PMCID: PMC9154234
- DOI: 10.1093/bioinformatics/btac298
Simulation and annotation of global acronyms
Abstract
Motivation: Global acronyms are used in written text without their formal definitions. This makes it difficult to automatically interpret their sense as acronyms tend to be ambiguous. Supervised machine learning approaches to sense disambiguation require large training datasets. In clinical applications, large datasets are difficult to obtain due to patient privacy. Manual data annotation creates an additional bottleneck.
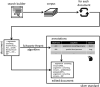
Results: We proposed an approach to automatically modifying scientific abstracts to (i) simulate global acronym usage and (ii) annotate their senses without the need for external sources or manual intervention. We implemented it as a web-based application, which can create large datasets that in turn can be used to train supervised approaches to word sense disambiguation of biomedical acronyms.
Availability and implementation: The datasets will be generated on demand based on a user query and will be downloadable from https://datainnovation.cardiff.ac.uk/acronyms/.
© The Author(s) 2022. Published by Oxford University Press.
References
-
- Agirre E., Stevenson M. (2006) Knowledge sources for WSD. In: Agirre, E. and Edmonds, P. (eds) Word Sense Disambiguation, pp. 217–251. https://doi.org/10.1007/978-1-4020-4809-8_8.
-
- Devlin J. et al. (2018) BERT: pre-training of deep bidirectional transformers for language understanding. In: Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Vol. 1, Association for Computational Linguistics, Minneapolis, Minnesota, pp. 4171–4186.