Long-read and chromosome-scale assembly of the hexaploid wheat genome achieves high resolution for research and breeding
- PMID: 35482491
- PMCID: PMC9049114
- DOI: 10.1093/gigascience/giac034
Long-read and chromosome-scale assembly of the hexaploid wheat genome achieves high resolution for research and breeding
Abstract
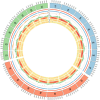
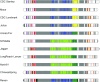
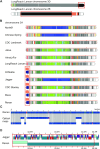
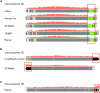
Background: The sequencing of the wheat (Triticum aestivum) genome has been a methodological challenge for many years owing to its large size (15.5 Gb), repeat content, and hexaploidy. Many initiatives aiming at obtaining a reference genome of cultivar Chinese Spring have been launched in the past years and it was achieved in 2018 as the result of a huge effort to combine short-read sequencing with many other resources. Reference-quality genome assemblies were then produced for other accessions, but the rapid evolution of sequencing technologies offers opportunities to reach high-quality standards at lower cost.
Results: Here, we report on an optimized procedure based on long reads produced on the Oxford Nanopore Technology PromethION device to assemble the genome of the French bread wheat cultivar Renan.
Conclusions: We provide the most contiguous chromosome-scale assembly of a bread wheat genome to date. Coupled with an annotation based on RNA-sequencing data, this resource will be valuable for the crop community and will facilitate the rapid selection of agronomically important traits. We also provide a framework to generate high-quality assemblies of complex genomes using ONT.
Keywords: genome assembly; haplotype characterization; hexaploid genome; introgressions; long-reads; nanopore sequencing; wheat.
© The Author(s) 2022. Published by Oxford University Press GigaScience.
Conflict of interest statement
J.M.A. received travel and accommodation expenses to speak at Oxford Nanopore Technologies conferences. J.M.A. and C.B. received accommodation expenses to speak at Bionano Genomics user meetings. The authors declare that they have no other competing interests.
Figures




References
-
- Marcussen T, Sandve SR, Heier L, et al. Ancient hybridizations among the ancestral genomes of bread wheat. Science. 2014;345(6194):1250092. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

