Activation of class 1 integron integrase is promoted in the intestinal environment
- PMID: 35482826
- PMCID: PMC9090394
- DOI: 10.1371/journal.pgen.1010177
Activation of class 1 integron integrase is promoted in the intestinal environment
Abstract
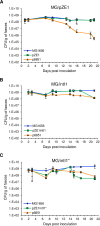
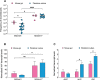
Class 1 integrons are widespread genetic elements playing a major role in the dissemination of antibiotic resistance. They allow bacteria to capture, express and exchange antibiotic resistance genes embedded within gene cassettes. Acquisition of gene cassettes is catalysed by the class 1 integron integrase, a site-specific recombinase playing a key role in the integron system. In in vitro planktonic culture, expression of intI1 is controlled by the SOS response, a regulatory network which mediates the repair of DNA damage caused by a wide range of bacterial stress, including antibiotics. However, in vitro experimental conditions are far from the real lifestyle of bacteria in natural environments such as the intestinal tract which is known to be a reservoir of integrons. In this study, we developed an in vivo model of intestinal colonization in gnotobiotic mice and used a recombination assay and quantitative real-time PCR, to investigate the induction of the SOS response and expression and activity of the class 1 integron integrase, IntI1. We found that the basal activity of IntI1 was higher in vivo than in vitro. In addition, we demonstrated that administration of a subinhibitory concentration of ciprofloxacin rapidly induced both the SOS response and intI1 expression that was correlated with an increase of the activity of IntI1. Our findings show that the gut is an environment in which the class 1 integron integrase is induced and active, and they highlight the potential role of integrons in the acquisition and/or expression of resistance genes in the gut, particularly during antibiotic therapy.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
The Stringent Response Promotes Antibiotic Resistance Dissemination by Regulating Integron Integrase Expression in Biofilms.mBio. 2016 Aug 16;7(4):e00868-16. doi: 10.1128/mBio.00868-16. mBio. 2016. PMID: 27531906 Free PMC article.
-
Off-Target Integron Activity Leads to Rapid Plasmid Compensatory Evolution in Response to Antibiotic Selection Pressure.mBio. 2023 Apr 25;14(2):e0253722. doi: 10.1128/mbio.02537-22. Epub 2023 Feb 22. mBio. 2023. PMID: 36840554 Free PMC article.
-
Diversity of Class 1 Integron Gene Cassette Rearrangements Selected under Antibiotic Pressure.J Bacteriol. 2015 Jul;197(13):2171-2178. doi: 10.1128/JB.02455-14. Epub 2015 Apr 20. J Bacteriol. 2015. PMID: 25897031 Free PMC article.
-
Using the class 1 integron-integrase gene as a proxy for anthropogenic pollution.ISME J. 2015 Jun;9(6):1269-79. doi: 10.1038/ismej.2014.226. Epub 2014 Dec 12. ISME J. 2015. PMID: 25500508 Free PMC article. Review.
-
Class 1 integrons, gene cassettes, mobility, and epidemiology.Eur J Clin Microbiol Infect Dis. 1999 Nov;18(11):761-70. doi: 10.1007/s100960050398. Eur J Clin Microbiol Infect Dis. 1999. PMID: 10614949 Review.
Cited by
-
Whole-Genome Sequencing of an Escherichia coli ST69 Strain Harboring blaCTX-M-27 on a Hybrid Plasmid.Infect Drug Resist. 2024 Feb 1;17:365-375. doi: 10.2147/IDR.S427571. eCollection 2024. Infect Drug Resist. 2024. PMID: 38318209 Free PMC article.
-
Genomic and Transcriptomic Analysis Reveal Multiple Strategies for the Cadmium Tolerance in Vibrio parahaemolyticus N10-18 Isolated from Aquatic Animal Ostrea gigas Thunberg.Foods. 2022 Nov 23;11(23):3777. doi: 10.3390/foods11233777. Foods. 2022. PMID: 36496584 Free PMC article.
-
Ongoing toxin-positive diphtheria outbreaks in a federal asylum centre in Switzerland, analysis July to September 2022.Euro Surveill. 2022 Nov;27(44):2200811. doi: 10.2807/1560-7917.ES.2022.27.44.2200811. Euro Surveill. 2022. PMID: 36330823 Free PMC article.
-
Characterization of antibiotic resistance genes and mobile genetic elements in Escherichia coli isolated from captive black bears.Sci Rep. 2024 Feb 2;14(1):2745. doi: 10.1038/s41598-024-52622-2. Sci Rep. 2024. PMID: 38302507 Free PMC article.
-
Biological and Synthetic Surfactants Increase Class I Integron Prevalence in Ex Situ Biofilms.Microorganisms. 2024 Mar 31;12(4):712. doi: 10.3390/microorganisms12040712. Microorganisms. 2024. PMID: 38674656 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

