Chemical Proteomics Reveals Off-Targets of the Anandamide Reuptake Inhibitor WOBE437
- PMID: 35482948
- PMCID: PMC9127799
- DOI: 10.1021/acschembio.2c00122
Chemical Proteomics Reveals Off-Targets of the Anandamide Reuptake Inhibitor WOBE437
Abstract
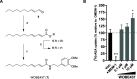
Anandamide or N-arachidonoylethanolamine (AEA) is a signaling lipid that modulates neurotransmitter release via activation of the type 1 cannabinoid receptor (CB1R) in the brain. Termination of anandamide signaling is thought to be mediated via a facilitated cellular reuptake process that utilizes a purported transporter protein. Recently, WOBE437 has been reported as a novel, natural product-based inhibitor of AEA reuptake that is active in cellular and in vivo models. To profile its target interaction landscape, we synthesized pac-WOBE, a photoactivatable probe derivative of WOBE437, and performed chemical proteomics in mouse neuroblastoma Neuro-2a cells. Surprisingly WOBE437, unlike the widely used selective inhibitor of AEA uptake OMDM-1, was found to increase AEA uptake in Neuro-2a cells. In line with this, WOBE437 reduced the cellular levels of AEA and related N-acylethanolamines (NAEs). Using pac-WOBE, we identified saccharopine dehydrogenase-like oxidoreductase (SCCPDH), vesicle amine transport 1 (VAT1), and ferrochelatase (FECH) as WOBE437-interacting proteins in Neuro-2a cells. Further genetic studies indicated that SCCPDH and VAT1 were not responsible for the WOBE437-induced reduction in NAE levels. Regardless of the precise mechanism of action of WOB437 in AEA transport, we have identified SSCPHD, VAT1, and FECH as unprecedented off-targets of this molecule which should be taken into account when interpreting its cellular and in vivo effects.
Conflict of interest statement
The authors declare no competing financial interest.
Figures





References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

