Real-time liver tumor localization via a single x-ray projection using deep graph neural network-assisted biomechanical modeling
- PMID: 35483350
- PMCID: PMC9233941
- DOI: 10.1088/1361-6560/ac6b7b
Real-time liver tumor localization via a single x-ray projection using deep graph neural network-assisted biomechanical modeling
Abstract
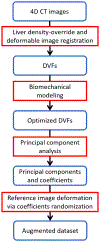
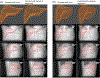
Objective.Real-time imaging is highly desirable in image-guided radiotherapy, as it provides instantaneous knowledge of patients' anatomy and motion during treatments and enables online treatment adaptation to achieve the highest tumor targeting accuracy. Due to extremely limited acquisition time, only one or few x-ray projections can be acquired for real-time imaging, which poses a substantial challenge to localize the tumor from the scarce projections. For liver radiotherapy, such a challenge is further exacerbated by the diminished contrast between the tumor and the surrounding normal liver tissues. Here, we propose a framework combining graph neural network-based deep learning and biomechanical modeling to track liver tumor in real-time from a single onboard x-ray projection.Approach.Liver tumor tracking is achieved in two steps. First, a deep learning network is developed to predict the liver surface deformation using image features learned from the x-ray projection. Second, the intra-liver deformation is estimated through biomechanical modeling, using the liver surface deformation as the boundary condition to solve tumor motion by finite element analysis. The accuracy of the proposed framework was evaluated using a dataset of 10 patients with liver cancer.Main results.The results show accurate liver surface registration from the graph neural network-based deep learning model, which translates into accurate, fiducial-less liver tumor localization after biomechanical modeling (<1.2 (±1.2) mm average localization error).Significance.The method demonstrates its potentiality towards intra-treatment and real-time 3D liver tumor monitoring and localization. It could be applied to facilitate 4D dose accumulation, multi-leaf collimator tracking and real-time plan adaptation. The method can be adapted to other anatomical sites as well.
Keywords: biomechanical modeling; deep learning; graph neural network; liver; real-time tumor localization; x-ray.
© 2022 Institute of Physics and Engineering in Medicine.
Figures


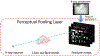
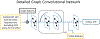




References
-
- Balter JM, Dawson LA, Kazanjian S, McGinn C, Brock KK, Lawrence T, and Ten Haken R. 2001. ‘Determination of ventilatory liver movement via radiographic evaluation of diaphragm position’, Int J Radiat Oncol Biol Phys, 51: 267–70. - PubMed
-
- Bianco S, Cadene R, Celona L, and Napoletano P. 2018. ‘Benchmark Analysis of Representative Deep Neural Network Architectures’, Ieee Access, 6: 64270–77.
-
- Booth J, Caillet V, Briggs A, Hardcastle N, Angelis G, Jayamanne D, Shepherd M, Podreka A, Szymura K, Nguyen DT, Poulsen P, O’Brien R, Harris B, Haddad C, Eade T, and Keall P. 2021. ‘MLC tracking for lung SABR is feasible, efficient and delivers high-precision target dose and lower normal tissue dose’, Radiotherapy and Oncology, 155: 131–37. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
