Analysis of mRNA vaccination-elicited RBD-specific memory B cells reveals strong but incomplete immune escape of the SARS-CoV-2 Omicron variant
- PMID: 35483354
- PMCID: PMC8986479
- DOI: 10.1016/j.immuni.2022.04.002
Analysis of mRNA vaccination-elicited RBD-specific memory B cells reveals strong but incomplete immune escape of the SARS-CoV-2 Omicron variant
Abstract
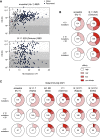
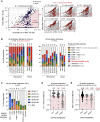
The SARS-CoV-2 Omicron variant can escape neutralization by vaccine-elicited and convalescent antibodies. Memory B cells (MBCs) represent another layer of protection against SARS-CoV-2, as they persist after infection and vaccination and improve their affinity. Whether MBCs elicited by mRNA vaccines can recognize the Omicron variant remains unclear. We assessed the affinity and neutralization potency against the Omicron variant of several hundred naturally expressed MBC-derived monoclonal IgG antibodies from vaccinated COVID-19-recovered and -naive individuals. Compared with other variants of concern, Omicron evaded recognition by a larger proportion of MBC-derived antibodies, with only 30% retaining high affinity against the Omicron RBD, and the reduction in neutralization potency was even more pronounced. Nonetheless, neutralizing MBC clones could be found in all the analyzed individuals. Therefore, despite the strong immune escape potential of the Omicron variant, these results suggest that the MBC repertoire generated by mRNA vaccines still provides some protection against the Omicron variant in vaccinated individuals.
Keywords: B cells; COVID-19; MBC; Omicron; SARS-CoV-2; VOC; affinity; germinal center; mRNA vaccine; memory B cells; variants.
Copyright © 2022. Published by Elsevier Inc.
Conflict of interest statement
Declaration of interests Outside of the submitted work, M. Mahévas received research funds from GSK and personal fees from LFB and Amgen. J.-C.W. received consulting fees from Institut Mérieux. P.B. received consulting fees from Regeneron Pharmaceuticals. J.-M.P. received personal fees from Abbvie, Gilead, Merck, and Siemens Healthcare. F.A.R. is a member of the board of MELETIOS Therapeutics and of the Scientific Advisory Board of eureKARE.
Figures



Comment in
-
Hide and seek with SARS-CoV-2: spike receptor-binding domain-specific memory B cells still recognize Omicron variant.Signal Transduct Target Ther. 2022 Oct 2;7(1):343. doi: 10.1038/s41392-022-01192-8. Signal Transduct Target Ther. 2022. PMID: 36184648 Free PMC article. No abstract available.
References
-
- Barnes C.O., West A.P., Huey-Tubman K.E., Hoffmann M.A.G., Sharaf N.G., Hoffman P.R., Koranda N., Gristick H.B., Gaebler C., Muecksch F., et al. Structures of human antibodies bound to SARS-CoV-2 spike reveal common epitopes and recurrent features of antibodies. Cell. 2020;182:828–842.e16. - PMC - PubMed
-
- Carreño J.M., Alshammary H., Tcheou J., Singh G., Raskin A.J., Kawabata H., Sominsky L.A., Clark J.J., Adelsberg D.C., Bielak D.A., et al. Activity of convalescent and vaccine serum against SARS-CoV-2 Omicron. Nature. 2022;602:682–688. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

