Immune cell infiltration pattern in non-small cell lung cancer PDX models is a model immanent feature and correlates with a distinct molecular and phenotypic make-up
- PMID: 35483746
- PMCID: PMC9052060
- DOI: 10.1136/jitc-2021-004412
Immune cell infiltration pattern in non-small cell lung cancer PDX models is a model immanent feature and correlates with a distinct molecular and phenotypic make-up
Abstract
Background: The field of cancer immunology is rapidly moving towards innovative therapeutic strategies, resulting in the need for robust and predictive preclinical platforms reflecting the immunological response to cancer. Well characterized preclinical models are essential for the development of predictive biomarkers in the oncology as well as the immune-oncology space. In the current study, gold standard preclinical models are being refined and combined with novel image analysis tools to meet those requirements.
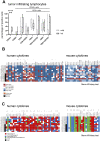
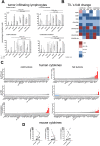
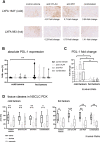
Methods: A panel of 14 non-small cell lung cancer patient-derived xenograft models (NSCLC PDX) was propagated in humanized NOD/Shi-scid/IL-2Rnull mice. The models were comprehensively characterized for relevant phenotypic and molecular features, including flow cytometry, immunohistochemistry, histology, whole exome sequencing and cytokine secretion.
Results: Models reflecting hot (>5% tumor-infiltrating lymphocytes/TILs) as opposed to cold tumors (<5% TILs) significantly differed regarding their cytokine profiles, molecular genetic aberrations, stroma content, and programmed cell death ligand-1 status. Treatment experiments including anti cytotoxic T-lymphocyte-associated protein 4, anti-programmed cell death 1 or the combination thereof across all 14 models in the single mouse trial format showed distinctive tumor growth response and spatial immune cell patterns as monitored by computerized analysis of digitized whole-slide images. Image analysis provided for the first time qualitative evaluation of the extent to which PDX models retain the histological features from their original human donors.
Conclusions: Deep phenotyping of PDX models in a humanized setting by combinations of computational pathology, immunohistochemistry, flow cytometry and proteomics enables the exhaustive analysis of innovative preclinical models and paves the way towards the development of translational biomarkers for immuno-oncology drugs.
Keywords: biomarkers, tumor; immunohistochemistry; lung neoplasms; tumor microenvironment.
© Author(s) (or their employer(s)) 2022. Re-use permitted under CC BY-NC. No commercial re-use. See rights and permissions. Published by BMJ.
Conflict of interest statement
Competing interests: None declared.
Figures




References
-
- Bertotti A, Migliardi G, Galimi F, et al. A molecularly annotated platform of patient-derived xenografts ("xenopatients") identifies HER2 as an effective therapeutic target in cetuximab-resistant colorectal cancer. Cancer Discov 2011;1:508–23. 10.1158/2159-8290.CD-11-0109 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
