A complete pedigree-based graph workflow for rare candidate variant analysis
- PMID: 35483961
- PMCID: PMC9104704
- DOI: 10.1101/gr.276387.121
A complete pedigree-based graph workflow for rare candidate variant analysis
Abstract
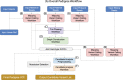
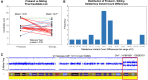
Methods that use a linear genome reference for genome sequencing data analysis are reference-biased. In the field of clinical genetics for rare diseases, a resulting reduction in genotyping accuracy in some regions has likely prevented the resolution of some cases. Pangenome graphs embed population variation into a reference structure. Although pangenome graphs have helped to reduce reference mapping bias, further performance improvements are possible. We introduce VG-Pedigree, a pedigree-aware workflow based on the pangenome-mapping tool of Giraffe and the variant calling tool DeepTrio using a specially trained model for Giraffe-based alignments. We demonstrate mapping and variant calling improvements in both single-nucleotide variants (SNVs) and insertion and deletion (indel) variants over those produced by alignments created using BWA-MEM to a linear-reference and Giraffe mapping to a pangenome graph containing data from the 1000 Genomes Project. We have also adapted and upgraded deleterious-variant (DV) detecting methods and programs into a streamlined workflow. We used these workflows in combination to detect small lists of candidate DVs among 15 family quartets and quintets of the Undiagnosed Diseases Program (UDP). All candidate DVs that were previously diagnosed using the Mendelian models covered by the previously published methods were recapitulated by these workflows. The results of these experiments indicate that a slightly greater absolute count of DVs are detected in the proband population than in their matched unaffected siblings.
© 2022 Markello et al.; Published by Cold Spring Harbor Laboratory Press.
Figures


References
-
- Clark MM, Stark Z, Farnaes L, Tan TY, White SM, Dimmock D, Kingsmore SF. 2018. Meta-analysis of the diagnostic and clinical utility of genome and exome sequencing and chromosomal microarray in children with suspected genetic diseases. NPJ Genom Med 3: 16. 10.1038/s41525-018-0053-8 - DOI - PMC - PubMed
