Kronos scRT: a uniform framework for single-cell replication timing analysis
- PMID: 35484127
- PMCID: PMC9050662
- DOI: 10.1038/s41467-022-30043-x
Kronos scRT: a uniform framework for single-cell replication timing analysis
Abstract
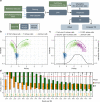
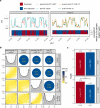
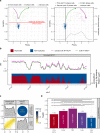
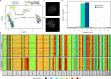
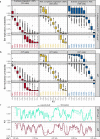
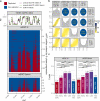
Mammalian genomes are replicated in a cell type-specific order and in coordination with transcription and chromatin organization. Currently, single-cell replication studies require individual processing of sorted cells, yielding a limited number (<100) of cells. Here, we develop Kronos scRT, a software for single-cell Replication Timing (scRT) analysis. Kronos scRT does not require a specific platform or cell sorting, which allows investigating large datasets obtained from asynchronous cells. By applying our tool to published data as well as droplet-based single-cell whole-genome sequencing data generated in this study, we exploit scRT from thousands of cells for different mouse and human cell lines. Our results demonstrate that although genomic regions are frequently replicated around their population average RT, replication can occur stochastically throughout S phase. Altogether, Kronos scRT allows fast and comprehensive investigations of the RT programme at the single-cell resolution for both homogeneous and heterogeneous cell populations.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Peace JM, Villwock SK, Zeytounian JL, Gan Y, Aparicio OM. Quantitative BrdU immunoprecipitation method demonstrates that Fkh1 and Fkh2 are rate-limiting activators of replication origins that reprogram replication timing in G1 phase. Genome Res. 2016;26:365–75. doi: 10.1101/gr.196857.115. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

