Molecular and functional heterogeneity in dorsal and ventral oligodendrocyte progenitor cells of the mouse forebrain in response to DNA damage
- PMID: 35484145
- PMCID: PMC9051058
- DOI: 10.1038/s41467-022-30010-6
Molecular and functional heterogeneity in dorsal and ventral oligodendrocyte progenitor cells of the mouse forebrain in response to DNA damage
Abstract
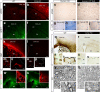
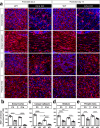
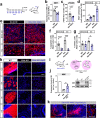
In the developing mouse forebrain, temporally distinct waves of oligodendrocyte progenitor cells (OPCs) arise from different germinal zones and eventually populate either dorsal or ventral regions, where they present as transcriptionally and functionally equivalent cells. Despite that, developmental heterogeneity influences adult OPC responses upon demyelination. Here we show that accumulation of DNA damage due to ablation of citron-kinase or cisplatin treatment cell-autonomously disrupts OPC fate, resulting in cell death and senescence in the dorsal and ventral subsets, respectively. Such alternative fates are associated with distinct developmental origins of OPCs, and with a different activation of NRF2-mediated anti-oxidant responses. These data indicate that, upon injury, dorsal and ventral OPC subsets show functional and molecular diversity that can make them differentially vulnerable to pathological conditions associated with DNA damage.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

