SARS-CoV-2-specific T-cell epitope repertoire in convalescent and mRNA-vaccinated individuals
- PMID: 35484232
- PMCID: PMC9064790
- DOI: 10.1038/s41564-022-01106-y
SARS-CoV-2-specific T-cell epitope repertoire in convalescent and mRNA-vaccinated individuals
Abstract
Continuously emerging variants of concern (VOCs) sustain the SARS-CoV-2 pandemic. The SARS-CoV-2 Omicron/B.1.1.529 VOC harbours multiple mutations in the spike protein associated with high infectivity and efficient evasion from humoral immunity induced by previous infection or vaccination. By performing in-depth comparisons of the SARS-CoV-2-specific T-cell epitope repertoire after infection and messenger RNA vaccination, we demonstrate that spike-derived epitopes were not dominantly targeted in convalescent individuals compared to non-spike epitopes. In vaccinees, however, we detected a broader spike-specific T-cell response compared to convalescent individuals. Booster vaccination increased the breadth of the spike-specific T-cell response in convalescent individuals but not in vaccinees with complete initial vaccination. In convalescent individuals and vaccinees, the targeted T-cell epitopes were broadly conserved between wild-type SARS-CoV-2 variant B and Omicron/B.1.1.529. Hence, our data emphasize the relevance of vaccine-induced spike-specific CD8+ T-cell responses in combating VOCs including Omicron/B.1.1.529 and support the benefit of boosting convalescent individuals with mRNA vaccines.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





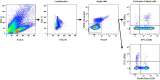
References
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

