Immunogenicity and therapeutic targeting of a public neoantigen derived from mutated PIK3CA
- PMID: 35484264
- PMCID: PMC9117146
- DOI: 10.1038/s41591-022-01786-3
Immunogenicity and therapeutic targeting of a public neoantigen derived from mutated PIK3CA
Abstract
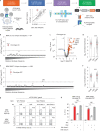
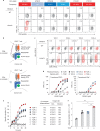
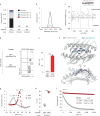
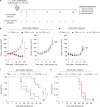
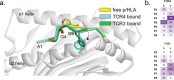
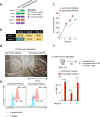
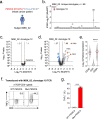
Public neoantigens (NeoAgs) represent an elite class of shared cancer-specific epitopes derived from recurrently mutated driver genes. Here we describe a high-throughput platform combining single-cell transcriptomic and T cell receptor (TCR) sequencing to establish whether mutant PIK3CA, among the most frequently genomically altered driver oncogenes, generates an immunogenic public NeoAg. Using this strategy, we developed a panel of TCRs that recognize an endogenously processed neopeptide encompassing a common PIK3CA hotspot mutation restricted by the prevalent human leukocyte antigen (HLA)-A*03:01 allele. Mechanistically, immunogenicity to this public NeoAg arises from enhanced neopeptide/HLA complex stability caused by a preferred HLA anchor substitution. Structural studies indicated that the HLA-bound neopeptide presents a comparatively 'featureless' surface dominated by the peptide's backbone. To bind this epitope with high specificity and affinity, we discovered that a lead TCR clinical candidate engages the neopeptide through an extended interface facilitated by an unusually long CDR3β loop. In patients with diverse malignancies, we observed NeoAg clonal conservation and spontaneous immunogenicity to the neoepitope. Finally, adoptive transfer of TCR-engineered T cells led to tumor regression in vivo in mice bearing PIK3CA-mutant tumors but not wild-type PIK3CA tumors. Together, these findings establish the immunogenicity and therapeutic potential of a mutant PIK3CA-derived public NeoAg.
© 2022. The Author(s).
Conflict of interest statement
S.S.C. and C.A.K. are inventors of the TCR discovery platform and the
Figures









References
-
- Parkhurst MR, et al. Unique neoantigens arise from somatic mutations in patients with gastrointestinal cancers. Cancer Discov. 2019;9:1022–1035. doi: 10.1158/2159-8290.CD-18-1494. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

