MYCN mediates cysteine addiction and sensitizes neuroblastoma to ferroptosis
- PMID: 35484422
- PMCID: PMC9050595
- DOI: 10.1038/s43018-022-00355-4
MYCN mediates cysteine addiction and sensitizes neuroblastoma to ferroptosis
Abstract
Aberrant expression of MYC transcription factor family members predicts poor clinical outcome in many human cancers. Oncogenic MYC profoundly alters metabolism and mediates an antioxidant response to maintain redox balance. Here we show that MYCN induces massive lipid peroxidation on depletion of cysteine, the rate-limiting amino acid for glutathione (GSH) biosynthesis, and sensitizes cells to ferroptosis, an oxidative, non-apoptotic and iron-dependent type of cell death. The high cysteine demand of MYCN-amplified childhood neuroblastoma is met by uptake and transsulfuration. When uptake is limited, cysteine usage for protein synthesis is maintained at the expense of GSH triggering ferroptosis and potentially contributing to spontaneous tumor regression in low-risk neuroblastomas. Pharmacological inhibition of both cystine uptake and transsulfuration combined with GPX4 inactivation resulted in tumor remission in an orthotopic MYCN-amplified neuroblastoma model. These findings provide a proof of concept of combining multiple ferroptosis targets as a promising therapeutic strategy for aggressive MYCN-amplified tumors.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures



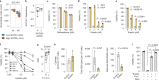









Comment in
-
Ferroptosis vulnerability in MYCN-driven neuroblastomas.Clin Transl Med. 2022 Aug;12(8):e963. doi: 10.1002/ctm2.963. Clin Transl Med. 2022. PMID: 35908258 Free PMC article. No abstract available.
References
-
- Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, metabolism, and cancer. Cancer Discov. 2015;5:1024–1039. doi: 10.1158/2159-8290.CD-15-0507. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

