Transcriptome and metabolome analyses reveal new insights into chlorophyll, photosynthesis, metal ion and phenylpropanoids related pathways during sugarcane ratoon chlorosis
- PMID: 35484490
- PMCID: PMC9052583
- DOI: 10.1186/s12870-022-03588-8
Transcriptome and metabolome analyses reveal new insights into chlorophyll, photosynthesis, metal ion and phenylpropanoids related pathways during sugarcane ratoon chlorosis
Abstract
Background: Ratoon sugarcane is susceptible to chlorosis, characterized by chlorophyll loss, poor growth, and a multitude of nutritional deficiency mainly occurring at young stage. Chlorosis would significantly reduce the cane production. The molecular mechanism underlying this phenomenon remains unknown. We analyzed the transcriptome and metabolome of chlorotic and non-chlorotic sugarcane leaves of the same age from the same field to gain molecular insights into this phenomenon.
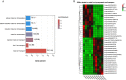
Results: The agronomic traits, such as plant height and the number of leaf, stalk node, and tillers declined in chlorotic sugarcane. Chlorotic leaves had substantially lower chlorophyll content than green leaves. A total of 11,776 differentially expressed genes (DEGs) were discovered in transcriptome analysis. In the KEGG enriched chlorophyll metabolism pathway, sixteen DEGs were found, eleven of which were down-regulated. Two photosynthesis pathways were also enriched with 32 genes downregulated and four genes up-regulated. Among the 81 enriched GO biological processes, there were four categories related to metal ion homeostasis and three related to metal ion transport. Approximately 400 metabolites were identified in metabolome analysis. The thirteen differentially expressed metabolites (DEMs) were all found down-regulated. The phenylpropanoid biosynthesis pathway was enriched in DEGs and DEMs, indicating a potentially vital role for phenylpropanoids in chlorosis.
Conclusions: Chlorophyll production, metal ion metabolism, photosynthesis, and some metabolites in the phenylpropanoid biosynthesis pathway were considerably altered in chlorotic ratoon sugarcane leaves. Our finding revealed the relation between chlorosis and these pathways, which will help expand our mechanistic understanding of ratoon sugarcane chlorosis.
Keywords: Chlorophyll metabolism; Metal ion metabolism; Phenylpropanoids biosynthesis; Photosynthesis; Ratoon sugarcane chlorosis.
© 2022. The Author(s).
Conflict of interest statement
The authors declared that this study was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures








References
-
- Zhang M, Govindaraju M. Sugarcane production in China. In: de Oliveira, A. o. , editor. Sugarcane - Technology and Research [Internet]. London: IntechOpen; 2018. 10.5772/intechopen.73113.
-
- Chen G-F, Liu Z, Huang Y-Y, Xiong L-M, Tan Y-M, Xing Y, Tang Q-Z. Factors responsible for sugarcane ratoon chlorosis in acid soil and its management in Guangxi province of China. Sugar Tech. 2016;18(5):500–504. doi: 10.1007/s12355-016-0425-2. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

