Deep learning from multiple experts improves identification of amyloid neuropathologies
- PMID: 35484610
- PMCID: PMC9052651
- DOI: 10.1186/s40478-022-01365-0
Deep learning from multiple experts improves identification of amyloid neuropathologies
Abstract
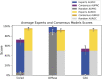
Pathologists can label pathologies differently, making it challenging to yield consistent assessments in the absence of one ground truth. To address this problem, we present a deep learning (DL) approach that draws on a cohort of experts, weighs each contribution, and is robust to noisy labels. We collected 100,495 annotations on 20,099 candidate amyloid beta neuropathologies (cerebral amyloid angiopathy (CAA), and cored and diffuse plaques) from three institutions, independently annotated by five experts. DL methods trained on a consensus-of-two strategy yielded 12.6-26% improvements by area under the precision recall curve (AUPRC) when compared to those that learned individualized annotations. This strategy surpassed individual-expert models, even when unfairly assessed on benchmarks favoring them. Moreover, ensembling over individual models was robust to hidden random annotators. In blind prospective tests of 52,555 subsequent expert-annotated images, the models labeled pathologies like their human counterparts (consensus model AUPRC = 0.74 cored; 0.69 CAA). This study demonstrates a means to combine multiple ground truths into a common-ground DL model that yields consistent diagnoses informed by multiple and potentially variable expert opinions.
Keywords: Algorithms; Amyloid beta; Consensus; Deep learning; Expert annotators; Histopathology.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures





References
-
- Campanella G, Hanna MG, Geneslaw L, Miraflor A, Werneck Krauss Silva V, Busam KJ, Brogi E, Reuter VE, Klimstra DS, Fuchs TJ. Clinical-grade computational pathology using weakly supervised deep learning on whole slide images. Nat Med. 2019;25:1301–1309. doi: 10.1038/s41591-019-0508-1. - DOI - PMC - PubMed
-
- Fillenbaum GG, van Belle G, Morris JC, Mohs RC, Mirra SS, Davis PC, Tariot PN, Silverman JM, Clark CM, Welsh-Bohmer KA, Heyman A. Consortium to Establish a Registry for Alzheimer’s Disease (CERAD): the first twenty years. Alzheimers Dement. 2008;4:96–109. doi: 10.1016/j.jalz.2007.08.005. - DOI - PMC - PubMed

