Molecular characterization and clinical outcome of B-cell precursor acute lymphoblastic leukemia with IG-MYC rearrangement
- PMID: 35484682
- PMCID: PMC9973471
- DOI: 10.3324/haematol.2021.280557
Molecular characterization and clinical outcome of B-cell precursor acute lymphoblastic leukemia with IG-MYC rearrangement
Abstract
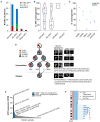
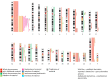
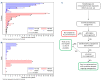
Rarely, immunophenotypically immature B-cell precursor acute lymphoblastic leukemia (BCP-ALL) carries an immunoglobulin- MYC rearrangement (IG-MYC-r). This can result in diagnostic confusion with Burkitt lymphoma/leukemia and use of individualized treatment schedules of unproven efficacy. Here we compare the molecular characteristics of these conditions and investigate historic clinical outcome data. We identified 90 cases registered in a national BCP-ALL clinical trial/registry. When present, diagnostic material underwent cytogenetic, exome, methylome and transcriptome analyses. The outcomes analyzed were 3-year event-free survival and overall survival. IG-MYC-r was identified in diverse cytogenetic backgrounds, co-existing with either established BCP-ALL-specific abnormalities (high hyperdiploidy, n=3; KMT2A-rearrangement, n=6; iAMP21, n=1; BCR-ABL1, n=1); BCL2/BCL6-rearrangements (n=15); or, most commonly, as the only defining feature (n=64). Within this final group, precursor-like V(D)J breakpoints predominated (8/9) and KRAS mutations were common (5/11). DNA methylation identified a cluster of V(D)J-rearranged cases, clearly distinct from Burkitt leukemia/lymphoma. Children with IG-MYC-r within that subgroup had a 3-year event-free survival of 47% and overall survival of 60%, representing a high-risk BCP-ALL. To develop effective management strategies this group of patients must be allowed access to contemporary, minimal residual disease-adapted, prospective clinical trial protocols.
Figures



Comment in
-
B-cell precursor leukemias with MYC-rearrangement come into the limelight.Haematologica. 2023 Mar 1;108(3):659-660. doi: 10.3324/haematol.2022.281112. Haematologica. 2023. PMID: 35484663 Free PMC article. No abstract available.
References
-
- Klapper W, Stoecklein H, Zeynalova S, et al. Structural aberrations affecting the MYC locus indicate a poor prognosis independent of clinical risk factors in diffuse large B-cell lymphomas treated within randomized trials of the German High-Grade Non-Hodgkin's Lymphoma Study Group (DSHNHL). Leukemia. 2008;22(12):2226-2229. - PubMed
-
- Savage KJ, Johnson NA, Ben-Neriah S, et al. MYC gene rearrangements are associated with a poor prognosis in diffuse large B-cell lymphoma patients treated with R-CHOP chemotherapy. Blood. 2009;114(17):3533-3537. - PubMed
-
- Ott G, Rosenwald A, Campo E. Understanding MYC-driven aggressive B-cell lymphomas: pathogenesis and classification. Blood. 2013;122(24):3884-3891. - PubMed
-
- Swerdlow SH, Campo E, Harris NL, et al. WHO Classification of Tumours of Haematopoietic and Lymphoid Tissues (Revised 4th edition): International Agency for Research on Cancer (IARC). 2017
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

