Interindividual Variability in Cytochrome P450 3A and 1A Activity Influences Sunitinib Metabolism and Bioactivation
- PMID: 35484684
- PMCID: PMC9131896
- DOI: 10.1021/acs.chemrestox.1c00426
Interindividual Variability in Cytochrome P450 3A and 1A Activity Influences Sunitinib Metabolism and Bioactivation
Abstract
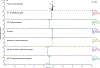
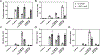
Sunitinib is an orally administered tyrosine kinase inhibitor associated with idiosyncratic hepatotoxicity; however, the mechanisms of this toxicity remain unclear. We have previously shown that cytochromes P450 1A2 and 3A4 catalyze sunitinib metabolic activation via oxidative defluorination leading to a chemically reactive, potentially toxic quinoneimine, trapped as a glutathione (GSH) conjugate (M5). The goals of this study were to determine the impact of interindividual variability in P450 1A and 3A activity on sunitinib bioactivation to the reactive quinoneimine and sunitinib N-dealkylation to the primary active metabolite N-desethylsunitinib (M1). Experiments were conducted in vitro using single-donor human liver microsomes and human hepatocytes. Relative sunitinib metabolite levels were measured by liquid chromatography-tandem mass spectrometry. In human liver microsomes, the P450 3A inhibitor ketoconazole significantly reduced M1 formation compared to the control. The P450 1A2 inhibitor furafylline significantly reduced defluorosunitinib (M3) and M5 formation compared to the control but had minimal effect on M1. In CYP3A5-genotyped human liver microsomes from 12 individual donors, M1 formation was highly correlated with P450 3A activity measured by midazolam 1'-hydroxylation, and M3 and M5 formation was correlated with P450 1A2 activity estimated by phenacetin O-deethylation. M3 and M5 formation was also associated with P450 3A5-selective activity. In sandwich-cultured human hepatocytes, the P450 3A inducer rifampicin significantly increased M1 levels. P450 1A induction by omeprazole markedly increased M3 formation and the generation of a quinoneimine-cysteine conjugate (M6) identified as a downstream metabolite of M5. The nonselective P450 inhibitor 1-aminobenzotriazole reduced each of these metabolites (M1, M3, and M6). Collectively, these findings indicate that P450 3A activity is a key determinant of sunitinib N-dealkylation to the active metabolite M1, and P450 1A (and potentially 3A5) activity influences sunitinib bioactivation to the reactive quinoneimine metabolite. Accordingly, modulation of P450 activity due to genetic and/or nongenetic factors may impact the risk of sunitinib-associated toxicities.
Conflict of interest statement
The authors declare no competing financial interest.
Figures




References
-
- Mendel DB; Laird AD; Xin X; Louie SG; Christensen JG; Li G; Schreck RE; Abrams TJ; Ngai TJ; Lee LB; Murray LJ; Carver J; Chan E; Moss KG; Haznedar JO; Sukbuntherng J; Blake RA; Sun L; Tang C; Miller T; Shirazian S; McMahon G; Cherrington JM In vivo antitumor activity of SU11248, a novel tyrosine kinase inhibitor targeting vascular endothelial growth factor and platelet-derived growth factor receptors: determination of a pharmacokinetic/pharmacodynamic relationship. Clin. Cancer Res 2003, 9, 327–337. - PubMed
-
- Abrams TJ; Lee LB; Murray LJ; Pryer NK; Cherrington JM SU11248 inhibits KIT and platelet-derived growth factor receptor beta in preclinical models of human small cell lung cancer. Mol. Cancer Ther 2003, 2, 471–478. - PubMed
-
- Goodman VL; Rock EP; Dagher R; Ramchandani RP; Abraham S; Gobburu JV; Booth BP; Verbois SL; Morse DE; Liang CY; Chidambaram N; Jiang JX; Tang S; Mahjoob K; Justice R; Pazdur R. Approval summary: sunitinib for the treatment of imatinib refractory or intolerant gastrointestinal stromal tumors and advanced renal cell carcinoma. Clin. Cancer Res 2007, 13, 1367–1373. - PubMed
-
- Chow LQ; Eckhardt SG Sunitinib: from rational design to clinical efficacy. J. Clin. Oncol 2007, 25, 884–896. - PubMed
-
- FDA. Sunitinib prescribing information, 2020. https://www.accessdata.fda.gov/drugsatfda_docs/label/2020/021938s037lbl.pdf.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

