Protein Flexibility and Dissociation Pathway Differentiation Can Explain Onset of Resistance Mutations in Kinases
- PMID: 35486370
- PMCID: PMC9256798
- DOI: 10.1002/anie.202200983
Protein Flexibility and Dissociation Pathway Differentiation Can Explain Onset of Resistance Mutations in Kinases
Abstract
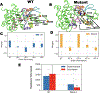
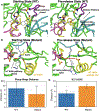
Understanding how mutations render a drug ineffective is a problem of immense relevance. Often the mechanism through which mutations cause drug resistance can be explained purely through thermodynamics. However, the more perplexing situation is when two proteins have the same drug binding affinities but different residence times. In this work, we demonstrate how all-atom molecular dynamics simulations using recent developments grounded in statistical mechanics can provide a detailed mechanistic rationale for such variances. We discover dissociation mechanisms for the anti-cancer drug Imatinib (Gleevec) against wild-type and the N368S mutant of Abl kinase. We show how this point mutation triggers far-reaching changes in the protein's flexibility and leads to a different, much faster, drug dissociation pathway. We believe that this work marks an efficient and scalable approach to obtain mechanistic insight into resistance mutations in biomolecular receptors that are hard to explain using a structural perspective.
Keywords: Kinase; Kinetics; Molecular Dynamics; Protein Flexibility; Resistance Mutations.
© 2022 Wiley-VCH GmbH.
Conflict of interest statement
Conflict of Interest
The authors declare no conflict of interest.
Figures
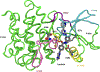


References
-
- Vogtherr M; Saxena K; Hoelder S; Grimme S; Betz M; Schieborr U; Pescatore B; Robin M; Delarbre L; Langer T, et al. Angewandte Chemie International Edition 2006, 45, 993–997. - PubMed
-
- Nielsen G; Jonker HR; Vajpai N; Grzesiek S; Schwalbe H ChemBioChem 2013, 14, 1799–1806. - PubMed
-
- Manning G; Whyte DB; Martinez R; Hunter T; Sudarsanam S Science 2002, 298, 1912–1934. - PubMed
-
- Roskoski R Pharmacological Research 2015, 100, 1–23. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

