Molecular basis of antibiotic self-resistance in a bee larvae pathogen
- PMID: 35487884
- PMCID: PMC9054821
- DOI: 10.1038/s41467-022-29829-w
Molecular basis of antibiotic self-resistance in a bee larvae pathogen
Abstract
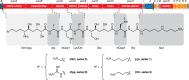
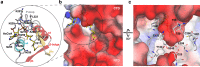
Paenibacillus larvae, the causative agent of the devastating honey-bee disease American Foulbrood, produces the cationic polyketide-peptide hybrid paenilamicin that displays antibacterial and antifungal activity. Its biosynthetic gene cluster contains a gene coding for the N-acetyltransferase PamZ. We show that PamZ acts as self-resistance factor in Paenibacillus larvae by deactivation of paenilamicin. Using tandem mass spectrometry, nuclear magnetic resonance spectroscopy and synthetic diastereomers, we identified the N-terminal amino group of the agmatinamic acid as the N-acetylation site. These findings highlight the pharmacophore region of paenilamicin, which we very recently identified as a ribosome inhibitor. Here, we further determined the crystal structure of PamZ:acetyl-CoA complex at 1.34 Å resolution. An unusual tandem-domain architecture provides a well-defined substrate-binding groove decorated with negatively-charged residues to specifically attract the cationic paenilamicin. Our results will help to understand the mode of action of paenilamicin and its role in pathogenicity of Paenibacillus larvae to fight American Foulbrood.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Lindström A, Korpela S, Fries I. Horizontal transmission of Paenibacillus larvae spores between honey bee (Apis mellifera) colonies through robbing. Apidologie. 2008;39:515–522. doi: 10.1051/apido:2008032. - DOI
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

