Alterations in microbiota of patients with COVID-19: potential mechanisms and therapeutic interventions
- PMID: 35487886
- PMCID: PMC9052735
- DOI: 10.1038/s41392-022-00986-0
Alterations in microbiota of patients with COVID-19: potential mechanisms and therapeutic interventions
Abstract
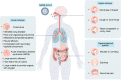
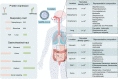
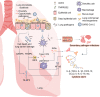
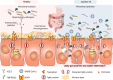
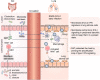
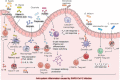
The global coronavirus disease 2019 (COVID-19) pandemic is currently ongoing. It is caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). A high proportion of COVID-19 patients exhibit gastrointestinal manifestations such as diarrhea, nausea, or vomiting. Moreover, the respiratory and gastrointestinal tracts are the primary habitats of human microbiota and targets for SARS-CoV-2 infection as they express angiotensin-converting enzyme-2 (ACE2) and transmembrane protease serine 2 (TMPRSS2) at high levels. There is accumulating evidence that the microbiota are significantly altered in patients with COVID-19 and post-acute COVID-19 syndrome (PACS). Microbiota are powerful immunomodulatory factors in various human diseases, such as diabetes, obesity, cancers, ulcerative colitis, Crohn's disease, and certain viral infections. In the present review, we explore the associations between host microbiota and COVID-19 in terms of their clinical relevance. Microbiota-derived metabolites or components are the main mediators of microbiota-host interactions that influence host immunity. Hence, we discuss the potential mechanisms by which microbiota-derived metabolites or components modulate the host immune responses to SARS-CoV-2 infection. Finally, we review and discuss a variety of possible microbiota-based prophylaxes and therapies for COVID-19 and PACS, including fecal microbiota transplantation (FMT), probiotics, prebiotics, microbiota-derived metabolites, and engineered symbiotic bacteria. This treatment strategy could modulate host microbiota and mitigate virus-induced inflammation.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures

References
-
- Round, J. L. & Palm, N. W. Causal effects of the microbiota on immune-mediated diseases. Sci Immunol. 3, eaao1603 (2018). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

