Comprehensive profiling of 1015 patients' exomes reveals genomic-clinical associations in colorectal cancer
- PMID: 35487942
- PMCID: PMC9055073
- DOI: 10.1038/s41467-022-30062-8
Comprehensive profiling of 1015 patients' exomes reveals genomic-clinical associations in colorectal cancer
Abstract
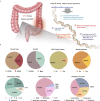
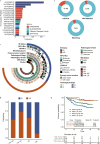
The genetic basis of colorectal cancer (CRC) and its clinical associations remain poorly understood due to limited samples or targeted genes in current studies. Here, we perform ultradeep whole-exome sequencing on 1015 patients with CRC as part of the ChangKang Project. We identify 46 high-confident significantly mutated genes, 8 of which mutate in 14.9% of patients: LYST, DAPK1, CR2, KIF16B, NPIPB15, SYTL2, ZNF91, and KIAA0586. With an unsupervised clustering algorithm, we propose a subtyping strategy that classisfies CRC patients into four genomic subtypes with distinct clinical characteristics, including hypermutated, chromosome instability with high risk, chromosome instability with low risk, and genome stability. Analysis of immunogenicity uncover the association of immunogenicity reduction with genomic subtypes and poor prognosis in CRC. Moreover, we find that mitochondrial DNA copy number is an independent factor for predicting the survival outcome of CRCs. Overall, our results provide CRC-related molecular features for clinical practice and a valuable resource for translational research.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





References
-
- Allison KH, Sledge GW. Heterogeneity and cancer. Oncology. 2014;28:772–778. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

