Life of double minutes: generation, maintenance, and elimination
- PMID: 35487993
- PMCID: PMC9470669
- DOI: 10.1007/s00412-022-00773-4
Life of double minutes: generation, maintenance, and elimination
Abstract
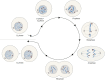
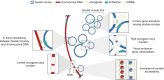
Advances in genome sequencing have revealed a type of extrachromosomal DNA, historically named double minutes (also referred to as ecDNA), to be common in a wide range of cancer types, but not in healthy tissues. These cancer-associated circular DNA molecules contain one or a few genes that are amplified when double minutes accumulate. Double minutes harbor oncogenes or drug resistance genes that contribute to tumor aggressiveness through copy number amplification in combination with favorable epigenetic properties. Unequal distribution of double minutes over daughter cells contributes to intratumoral heterogeneity, thereby increasing tumor adaptability. In this review, we discuss various models delineating the mechanism of generation of double minutes. Furthermore, we highlight how double minutes are maintained, how they evolve, and discuss possible mechanisms driving their elimination.
Keywords: Double minutes; Extrachromosomal DNA; Extrachromosomal oncogene amplification; Gene amplification; ecDNA.
© 2022. The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




References
-
- Alitalo K, Schwab M, Lin CC, et al. Homogeneously staining chromosomal regions contain amplified copies of an abundantly expressed cellular oncogene (c-myc) in malignant neuroendocrine cells from a human colon carcinoma. Proc Natl Acad Sci U S A. 1983;80:1707–1711. doi: 10.1073/pnas.80.6.1707. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

