Integrated analysis of single-cell and bulk RNA sequencing data reveals a pan-cancer stemness signature predicting immunotherapy response
- PMID: 35488273
- PMCID: PMC9052621
- DOI: 10.1186/s13073-022-01050-w
Integrated analysis of single-cell and bulk RNA sequencing data reveals a pan-cancer stemness signature predicting immunotherapy response
Abstract
Background: Although immune checkpoint inhibitor (ICI) is regarded as a breakthrough in cancer therapy, only a limited fraction of patients benefit from it. Cancer stemness can be the potential culprit in ICI resistance, but direct clinical evidence is lacking.
Methods: Publicly available scRNA-Seq datasets derived from ICI-treated patients were collected and analyzed to elucidate the association between cancer stemness and ICI response. A novel stemness signature (Stem.Sig) was developed and validated using large-scale pan-cancer data, including 34 scRNA-Seq datasets, The Cancer Genome Atlas (TCGA) pan-cancer cohort, and 10 ICI transcriptomic cohorts. The therapeutic value of Stem.Sig genes was further explored using 17 CRISPR datasets that screened potential immunotherapy targets.
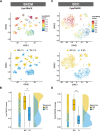
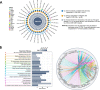
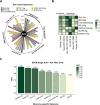
Results: Cancer stemness, as evaluated by CytoTRACE, was found to be significantly associated with ICI resistance in melanoma and basal cell carcinoma (both P < 0.001). Significantly negative association was found between Stem.Sig and anti-tumor immunity, while positive correlations were detected between Stem.Sig and intra-tumoral heterogenicity (ITH) / total mutational burden (TMB). Based on this signature, machine learning model predicted ICI response with an AUC of 0.71 in both validation and testing set. Remarkably, compared with previous well-established signatures, Stem.Sig achieved better predictive performance across multiple cancers. Moreover, we generated a gene list ranked by the average effect of each gene to enhance tumor immune response after genetic knockout across different CRISPR datasets. Then we matched Stem.Sig to this gene list and found Stem.Sig significantly enriched 3% top-ranked genes from the list (P = 0.03), including EMC3, BECN1, VPS35, PCBP2, VPS29, PSMF1, GCLC, KXD1, SPRR1B, PTMA, YBX1, CYP27B1, NACA, PPP1CA, TCEB2, PIGC, NR0B2, PEX13, SERF2, and ZBTB43, which were potential therapeutic targets.
Conclusions: We revealed a robust link between cancer stemness and immunotherapy resistance and developed a promising signature, Stem.Sig, which showed increased performance in comparison to other signatures regarding ICI response prediction. This signature could serve as a competitive tool for patient selection of immunotherapy. Meanwhile, our study potentially paves the way for overcoming immune resistance by targeting stemness-associated genes.
Keywords: Big data analysis; Immune checkpoint therapy; Pan-cancer; Single-cell sequencing; Stemness.
© 2022. The Author(s).
Conflict of interest statement
The authors declare that they have no competing interests.
Figures



References
-
- Sharma P, Siddiqui BA, Anandhan S, Yadav SS, Subudhi SK, Gao J, et al. The Next Decade of Immune Checkpoint Therapy. Cancer Discov. 2021;11:838–857. doi: 10.1158/2159-8290.CD-20-1680. - DOI - PubMed
-
- Hakimi AA, Voss MH, Kuo F, Sanchez A, Liu M, Nixon BG, et al. Transcriptomic Profiling of the Tumor Microenvironment Reveals Distinct Subgroups of Clear Cell Renal Cell Cancer: Data from a Randomized Phase III Trial. Cancer Discov. 2019;9:510–525. doi: 10.1158/2159-8290.CD-18-0957. - DOI - PMC - PubMed
-
- Ott PA, Bang Y-J, Piha-Paul SA, Razak ARA, Bennouna J, Soria J-C, et al. T-Cell–Inflamed Gene-Expression Profile, Programmed Death Ligand 1 Expression, and Tumor Mutational Burden Predict Efficacy in Patients Treated With Pembrolizumab Across 20 Cancers: KEYNOTE-028. J Clin Oncol. 2019;37:318–327. doi: 10.1200/JCO.2018.78.2276. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

