Nurr1 Modulation Mediates Neuroprotective Effects of Statins
- PMID: 35488520
- PMCID: PMC9218776
- DOI: 10.1002/advs.202104640
Nurr1 Modulation Mediates Neuroprotective Effects of Statins
Abstract
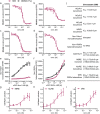
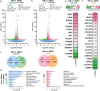
The ligand-sensing transcription factor Nurr1 emerges as a promising therapeutic target for neurodegenerative pathologies but Nurr1 ligands for functional studies and therapeutic validation are lacking. Here pronounced Nurr1 modulation by statins for which clinically relevant neuroprotective effects are demonstrated, is reported. Several statins directly affect Nurr1 activity in cellular and cell-free settings with low micromolar to sub-micromolar potencies. Simvastatin as example exhibits anti-inflammatory effects in astrocytes, which are abrogated by Nurr1 knockdown. Differential gene expression analysis in native and Nurr1-silenced cells reveals strong proinflammatory effects of Nurr1 knockdown while simvastatin treatment induces several neuroprotective mechanisms via Nurr1 involving changes in inflammatory, metabolic and cell cycle gene expression. Further in vitro evaluation confirms reduced inflammatory response, improved glucose metabolism, and cell cycle inhibition of simvastatin-treated neuronal cells. These findings suggest Nurr1 involvement in the well-documented but mechanistically elusive neuroprotection by statins.
Keywords: Alzheimer's disease; NR4A2; Parkinson's disease; multiple sclerosis; nuclear receptor related-1.
© 2022 The Authors. Advanced Science published by Wiley-VCH GmbH.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Similar articles
-
Molecular Insights into NR4A2(Nurr1): an Emerging Target for Neuroprotective Therapy Against Neuroinflammation and Neuronal Cell Death.Mol Neurobiol. 2019 Aug;56(8):5799-5814. doi: 10.1007/s12035-019-1487-4. Epub 2019 Jan 25. Mol Neurobiol. 2019. PMID: 30684217 Review.
-
Hypoxia-inducible factor 1 alpha and nuclear-related receptor 1 as targets for neuroprotection by albendazole in a rat rotenone model of Parkinson's disease.Clin Exp Pharmacol Physiol. 2019 Dec;46(12):1141-1150. doi: 10.1111/1440-1681.13162. Epub 2019 Sep 9. Clin Exp Pharmacol Physiol. 2019. PMID: 31408200
-
Nurr1 and Retinoid X Receptor Ligands Stimulate Ret Signaling in Dopamine Neurons and Can Alleviate α-Synuclein Disrupted Gene Expression.J Neurosci. 2015 Oct 21;35(42):14370-85. doi: 10.1523/JNEUROSCI.1155-15.2015. J Neurosci. 2015. PMID: 26490873 Free PMC article.
-
A Nurr1 Agonist Derived from the Natural Ligand DHI Induces Neuroprotective Gene Expression.J Med Chem. 2025 Feb 27;68(4):4829-4847. doi: 10.1021/acs.jmedchem.4c03104. Epub 2025 Feb 7. J Med Chem. 2025. PMID: 39919139 Free PMC article.
-
Nurr1-Based Therapies for Parkinson's Disease.CNS Neurosci Ther. 2016 May;22(5):351-9. doi: 10.1111/cns.12536. Epub 2016 Mar 25. CNS Neurosci Ther. 2016. PMID: 27012974 Free PMC article. Review.
Cited by
-
Neurodegenerative Diseases: Unraveling the Heterogeneity of Astrocytes.Cells. 2024 May 27;13(11):921. doi: 10.3390/cells13110921. Cells. 2024. PMID: 38891053 Free PMC article. Review.
-
Orphan nuclear receptor transcription factors as drug targets.Transcription. 2025 Apr-Jun;16(2-3):224-260. doi: 10.1080/21541264.2025.2521766. Epub 2025 Jul 11. Transcription. 2025. PMID: 40646688 Free PMC article. Review.
-
Development of Nurr1 agonists from amodiaquine by scaffold hopping and fragment growing.Commun Chem. 2024 Jun 29;7(1):149. doi: 10.1038/s42004-024-01224-0. Commun Chem. 2024. PMID: 38951694 Free PMC article.
-
Carboxylic Acid Bioisosteres Boost Nurr1 Agonist Selectivity.J Med Chem. 2025 Aug 14;68(15):16212-16226. doi: 10.1021/acs.jmedchem.5c01140. Epub 2025 Jul 17. J Med Chem. 2025. PMID: 40674328 Free PMC article.
-
Structure-Guided Design of Nurr1 Agonists Derived from the Natural Ligand Dihydroxyindole.J Med Chem. 2023 Oct 12;66(19):13556-13567. doi: 10.1021/acs.jmedchem.3c00852. Epub 2023 Sep 26. J Med Chem. 2023. PMID: 37751901 Free PMC article.
References
-
- Wang Z., Benoit G., Liu J., Prasad S., Aarnisalo P., Liu X., Xu H., Walker N. P. C., Perlmann T., Nature 2003, 423, 555. - PubMed
-
- Decressac M., Volakakis N., Björklund A., Perlmann T., Nat. Rev. Neurol. 2013, 9, 629. - PubMed
-
- Liu W., Gao Y., Chang N., Biochem. Biophys. Res. Commun. 2017, 482, 1312. - PubMed
-
- Montarolo F., Perga S., Martire S., Bertolotto A., Inflammation Res. 2015, 64, 841. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
