Paired nicking-mediated COL17A1 reframing for junctional epidermolysis bullosa
- PMID: 35490295
- PMCID: PMC9372311
- DOI: 10.1016/j.ymthe.2022.04.020
Paired nicking-mediated COL17A1 reframing for junctional epidermolysis bullosa
Abstract
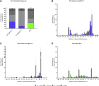
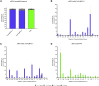
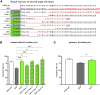
Junctional epidermolysis bullosa (JEB) is a debilitating hereditary skin disorder caused by mutations in genes encoding laminin-332, type XVII collagen (C17), and integrin-α6β4, which maintain stability between the dermis and epidermis. We designed patient-specific Cas9-nuclease- and -nickase-based targeting strategies for reframing a common homozygous deletion in exon 52 of COL17A1 associated with a lack of full-length C17 expression. Subsequent characterization of protein restoration, indel composition, and divergence of DNA and mRNA outcomes after treatment revealed auspicious efficiency, safety, and precision profiles for paired nicking-based COL17A1 editing. Almost 46% of treated primary JEB keratinocytes expressed reframed C17. Reframed COL17A1 transcripts predominantly featured 25- and 37-nt deletions, accounting for >42% of all edits and encoding C17 protein variants that localized accurately to the cell membrane. Furthermore, corrected cells showed accurate shedding of the extracellular 120-kDa C17 domain and improved adhesion capabilities to laminin-332 compared with untreated JEB cells. Three-dimensional (3D) skin equivalents demonstrated accurate and continuous deposition of C17 within the basal membrane zone between epidermis and dermis. Our findings constitute, for the first time, gene-editing-based correction of a COL17A1 mutation and demonstrate the superiority of proximal paired nicking strategies based on Cas9 D10A nickase over wild-type Cas9-based strategies for gene reframing in a clinical context.
Keywords: CRISPR-Cas9; JEB; gene reframing; junctional epidermolysis bullosa; next-generation sequencing; paired nicking; skin equivalents.
Copyright © 2022 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests T.C. and S.A.H. have filed a patent application for Abnoba-Seq. T.C. has a sponsored research collaboration with Cellectis and is an advisor to Cimeo Therapeutics and Excision BioTherapeutics. The other authors declare no competing interests.
Figures




References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

