Prospects for probiotics in social bees
- PMID: 35491599
- PMCID: PMC9058534
- DOI: 10.1098/rstb.2021.0156
Prospects for probiotics in social bees
Abstract
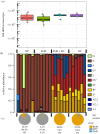
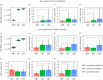
Social corbiculate bees are major pollinators. They have characteristic bacterial microbiomes associated with their hives and their guts. In honeybees and bumblebees, worker guts contain a microbiome composed of distinctive bacterial taxa shown to benefit hosts. These benefits include stimulating immune and metabolic pathways, digesting or detoxifying food, and defending against pathogens and parasites. Stressors including toxins and poor nutrition disrupt the microbiome and increase susceptibility to opportunistic pathogens. Administering probiotic bacterial strains may improve the health of individual bees and of hives, and several commercial probiotics are available for bees. However, evidence for probiotic benefits is lacking or mixed. Most bacterial species used in commercial probiotics are not native to bee guts. We present new experimental results showing that cultured strains of native bee gut bacteria colonize robustly while bacteria in a commercial probiotic do not establish in bee guts. A defined community of native bee gut bacteria resembles unperturbed native gut communities in its activation of genes for immunity and metabolism in worker bees. Although many questions remain unanswered, the development of natural probiotics for honeybees, or for commercially managed bumblebees, is a promising direction for protecting the health of managed bee colonies. This article is part of the theme issue 'Natural processes influencing pollinator health: from chemistry to landscapes'.
Keywords: Gilliamella; Nosema; Snodgrassella; apiculture; foulbrood; microbiome engraftment.
Conflict of interest statement
S.P.L., J.E.P., and N.A.M. are authors on a United States patent application involving use of native bee gut bacteria as probiotics to improve bee health.
Figures

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

