Anti-SARS-CoV-2 activities of tanshinone IIA, carnosic acid, rosmarinic acid, salvianolic acid, baicalein, and glycyrrhetinic acid between computational and in vitro insights
- PMID: 35492070
- PMCID: PMC9040650
- DOI: 10.1039/d1ra05268c
Anti-SARS-CoV-2 activities of tanshinone IIA, carnosic acid, rosmarinic acid, salvianolic acid, baicalein, and glycyrrhetinic acid between computational and in vitro insights
Abstract
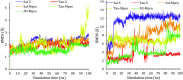
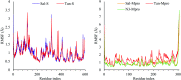
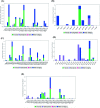
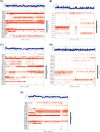
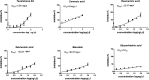
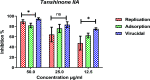
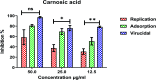
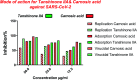
Six compounds namely, tanshinone IIA (1), carnosic acid (2), rosmarinic acid (3), salvianolic acid B (4), baicalein (5), and glycyrrhetinic acid (6) were screened for their anti-SARS-CoV-2 activities against both the spike (S) and main protease (Mpro) receptors using molecular docking studies. Molecular docking recommended the superior affinities of both salvianolic acid B (4) and glycyrrhetinic acid (6) as the common results from the previously published computational articles. On the other hand, their actual anti-SARS-CoV-2 activities were tested in vitro using plaque reduction assay to calculate their IC50 values after measuring their CC50 values using MTT assay on Vero E6 cells. Surprisingly, tanshinone IIA (1) was the most promising member with IC50 equals 4.08 ng μl-1. Also, both carnosic acid (2) and rosmarinic acid (3) showed promising IC50 values of 15.37 and 25.47 ng μl-1, respectively. However, salvianolic acid (4) showed a weak anti-SARS-CoV-2 activity with an IC50 value equals 58.29 ng μl-1. Furthermore, molecular dynamics simulations for 100 ns were performed for the most active compound from the computational point of view (salvianolic acid 4), besides, the most active one biologically (tanshinone IIA 1) on both the S and Mpro complexes of them (four different molecular dynamics processes) to confirm the docking results and give more insights regarding the stability of both compounds inside the SARS-CoV-2 mentioned receptors, respectively. Also, to understand the mechanism of action for the tested compounds towards SARS-CoV-2 inhibition it was necessary to examine the mode of action for the most two promising compounds, tanshinone IIA (1) and carnosic acid (2). Both compounds (1 and 2) showed very promising virucidal activity with a most prominent inhibitory effect on viral adsorption rather than its replication. This recommended the predicted activity of the two compounds against the S protein of SARS-CoV-2 rather than its Mpro protein. Our results could be very promising to rearrange the previously mentioned compounds based on their actual inhibitory activities towards SARS-CoV-2 and to search for the reasons behind the great differences between their in silico and in vitro results against SARS-CoV-2. Finally, we recommend further advanced preclinical and clinical studies especially for tanshinone IIA (1) to be rapidly applied in COVID-19 management either alone or in combination with carnosic acid (2), rosmarinic acid (3), and/or salvianolic acid (4).
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
In vitro and computational insights revealing the potential inhibitory effect of Tanshinone IIA against influenza A virus.Comput Biol Med. 2022 Feb;141:105149. doi: 10.1016/j.compbiomed.2021.105149. Epub 2021 Dec 17. Comput Biol Med. 2022. PMID: 34953359
-
In a search for potential drug candidates for combating COVID-19: computational study revealed salvianolic acid B as a potential therapeutic targeting 3CLpro and spike proteins.J Biomol Struct Dyn. 2022;40(19):8866-8893. doi: 10.1080/07391102.2021.1918256. Epub 2021 Apr 30. J Biomol Struct Dyn. 2022. PMID: 33928870
-
Design and statistical optimisation of emulsomal nanoparticles for improved anti-SARS-CoV-2 activity of N-(5-nitrothiazol-2-yl)-carboxamido candidates: in vitro and in silico studies.J Enzyme Inhib Med Chem. 2023 Dec;38(1):2202357. doi: 10.1080/14756366.2023.2202357. J Enzyme Inhib Med Chem. 2023. PMID: 37092260 Free PMC article.
-
Computational molecular docking and virtual screening revealed promising SARS-CoV-2 drugs.Precis Clin Med. 2021 Jan 18;4(1):1-16. doi: 10.1093/pcmedi/pbab001. eCollection 2021 Mar. Precis Clin Med. 2021. PMID: 33842834 Free PMC article. Review.
-
Natural Agents Modulating ACE-2: A Review of Compounds with Potential against SARS-CoV-2 Infections.Curr Pharm Des. 2021;27(13):1588-1596. doi: 10.2174/1381612827666210114150607. Curr Pharm Des. 2021. PMID: 33459225 Review.
Cited by
-
Protein structure-based in-silico approaches to drug discovery: Guide to COVID-19 therapeutics.Mol Aspects Med. 2023 Jun;91:101151. doi: 10.1016/j.mam.2022.101151. Epub 2022 Oct 28. Mol Aspects Med. 2023. PMID: 36371228 Free PMC article. Review.
-
Identification of Leading Compounds from Euphorbia neriifolia (Dudsor) Extracts as a Potential Inhibitor of SARS-CoV-2 ACE2-RBDS1 Receptor Complex: An Insight from Molecular Docking ADMET Profiling and MD-simulation Studies.Euroasian J Hepatogastroenterol. 2023 Jul-Dec;13(2):89-107. doi: 10.5005/jp-journals-10018-1414. Euroasian J Hepatogastroenterol. 2023. PMID: 38222948 Free PMC article.
-
Exploring the Antiviral Potential of Esters of Cinnamic Acids with Quercetin.Viruses. 2024 Apr 24;16(5):665. doi: 10.3390/v16050665. Viruses. 2024. PMID: 38793547 Free PMC article.
-
Identification of sulphonamide-tethered N-((triazol-4-yl)methyl)isatin derivatives as inhibitors of SARS-CoV-2 main protease.J Enzyme Inhib Med Chem. 2023 Dec;38(1):2234665. doi: 10.1080/14756366.2023.2234665. J Enzyme Inhib Med Chem. 2023. PMID: 37434404 Free PMC article.
-
A review of typical biological activities of glycyrrhetinic acid and its derivatives.RSC Adv. 2024 Feb 22;14(10):6557-6597. doi: 10.1039/d3ra08025k. eCollection 2024 Feb 21. RSC Adv. 2024. PMID: 38390501 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Miscellaneous

