A network-based approach to identify expression modules underlying rejection in pediatric liver transplantation
- PMID: 35492246
- PMCID: PMC9044102
- DOI: 10.1016/j.xcrm.2022.100605
A network-based approach to identify expression modules underlying rejection in pediatric liver transplantation
Abstract
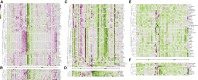
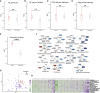
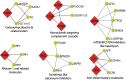
Selecting the right immunosuppressant to ensure rejection-free outcomes poses unique challenges in pediatric liver transplant (LT) recipients. A molecular predictor can comprehensively address these challenges. Currently, there are no well-validated blood-based biomarkers for pediatric LT recipients before or after LT. Here, we discover and validate separate pre- and post-LT transcriptomic signatures of rejection. Using an integrative machine learning approach, we combine transcriptomics data with the reference high-quality human protein interactome to identify network module signatures, which underlie rejection. Unlike gene signatures, our approach is inherently multivariate and more robust to replication and captures the structure of the underlying network, encapsulating additive effects. We also identify, in an individual-specific manner, signatures that can be targeted by current anti-rejection drugs and other drugs that can be repurposed. Our approach can enable personalized adjustment of drug regimens for the dominant targetable pathways before and after LT in children.
Keywords: liver transplantation; molecular diagnostics; network systems biology; pediatric samples; rejection; systems immunology.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Current Practice in Immunosuppression in Pediatric Liver Transplantation.Curr Pharm Des. 2020;26(28):3402-3405. doi: 10.2174/1381612826666200614181526. Curr Pharm Des. 2020. PMID: 32534559
-
Evaluating non-adherence to immunosuppressant medications in pediatric liver transplant recipients.Pediatr Transplant. 2008 May;12(3):284-8. doi: 10.1111/j.1399-3046.2008.00923.x. Epub 2008 Mar 4. Pediatr Transplant. 2008. PMID: 18331387
-
The absence of chronic rejection in pediatric primary liver transplant patients who are maintained on tacrolimus-based immunosuppression: a long-term analysis.Transplantation. 2003 Apr 15;75(7):1020-5. doi: 10.1097/01.TP.0000056168.79903.20. Transplantation. 2003. PMID: 12698091
-
Calcineurin inhibitor-free immunosuppression in pediatric renal transplantation: a viable option?Paediatr Drugs. 2011 Feb 1;13(1):49-69. doi: 10.2165/11538530-000000000-00000. Paediatr Drugs. 2011. PMID: 21162600 Review.
-
Liver transplantation. The pediatric challenge.Clin Liver Dis. 2000 Nov;4(4):879-927. doi: 10.1016/s1089-3261(05)70146-x. Clin Liver Dis. 2000. PMID: 11232362 Review.
Cited by
-
Prediction of the risk of transplant rejection based on RNA sequencing data of PBMCs before transplantation.Sci Rep. 2025 Aug 4;15(1):28391. doi: 10.1038/s41598-025-09780-8. Sci Rep. 2025. PMID: 40759993 Free PMC article.
-
Artificial Intelligence in Pediatric Liver Transplantation: Opportunities and Challenges of a New Era.Children (Basel). 2024 Aug 15;11(8):996. doi: 10.3390/children11080996. Children (Basel). 2024. PMID: 39201931 Free PMC article. Review.
-
Artificial intelligence applied to 'omics data in liver disease: towards a personalised approach for diagnosis, prognosis and treatment.Gut. 2025 Jan 17;74(2):295-311. doi: 10.1136/gutjnl-2023-331740. Gut. 2025. PMID: 39174307 Free PMC article. Review.
-
The Role of Dynamic DNA Methylation in Liver Transplant Rejection in Children.Transplant Direct. 2022 Oct 14;8(11):e1394. doi: 10.1097/TXD.0000000000001394. eCollection 2022 Nov. Transplant Direct. 2022. PMID: 36259078 Free PMC article.
-
From bench to bedside via bytes: Multi-omic immunoprofiling and integration using machine learning and network approaches.Hum Vaccin Immunother. 2023 Dec 15;19(3):2282803. doi: 10.1080/21645515.2023.2282803. Epub 2023 Dec 15. Hum Vaccin Immunother. 2023. PMID: 38100557 Free PMC article. Review.
References
-
- Waki K., Sugawara Y., Mizuta K., Fujita H., Kadowaki T., Kokudo N. Living-donor liver transplantation at the University of Tokyo, 1996-2011: the impact of HLA matching and a positive crossmatch on long-term survival and tolerance. Clin. Transpl. 2011:223–235. - PubMed
-
- Jain A., Demetris A.J., Kashyap R., Blakomer K., Ruppert K., Khan A., Rohal S., Starzl T.E., Fung J.J. Does tacrolimus offer virtual freedom from chronic rejection after primary liver transplantation? Risk and prognostic factors in 1,048 liver transplantations with a mean follow-up of 6 years. Liver Transpl. 2001;7:623–630. doi: 10.1053/jlts.2001.25364. - DOI - PMC - PubMed
-
- Ashokkumar C., Talukdar A., Sun Q., Higgs B.W., Janosky J., Wilson P., Mazariegos G., Jaffe R., Demetris A., Dobberstein J., et al. Allospecific CD154+ T cells associate with rejection risk after pediatric liver transplantation. Am. J. Transpl. 2009;9:179–191. doi: 10.1111/j.1600-6143.2008.02459.x. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases

