Identifying the specific-targeted marine cerebrosides against SARS-CoV-2: an integrated computational approach
- PMID: 35492761
- PMCID: PMC9043436
- DOI: 10.1039/d1ra07103c
Identifying the specific-targeted marine cerebrosides against SARS-CoV-2: an integrated computational approach
Abstract
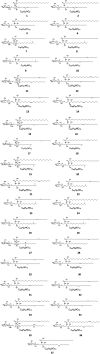
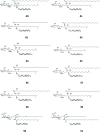
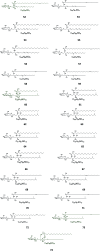
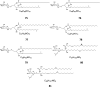
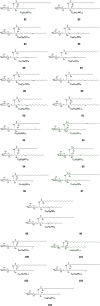
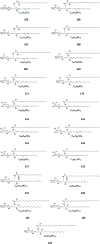
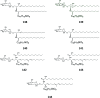
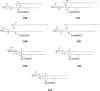
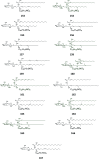
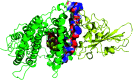
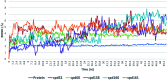
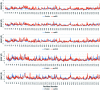
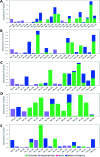
Cerebrosides are a group of metabolites belonging to the glycosphingolipids class of natural products. So far, 167 cerebrosides, compounds 1-167, have been isolated from diverse marine organisms or microorganisms. The as yet smaller number of compounds that have been studied more in depth proves a potential against challenging diseases, such as cancer, a range of viral and bacterial diseases, as well as inflammation. This review provides a comprehensive summary on this so far under-explored class of compounds, their chemical structures, bioactivities, and their marine sources, with a full coverage to the end of 2020. Today, the global pandemic concern, COVID-19, has claimed millions of death cases around the world, making the development of anti-SARS-CoV-2 drugs urgently needed for such a battle. Accordingly, selected examples from all subclasses of cerebrosides were virtually screened for potential inhibition of SARS-CoV-2 proteins that are crucially involved in the viral-host interaction, viral replication, or in disease progression. The results highlight five cerebrosides that could preferentially bind to the hACE2 protein, with binding scores between -7.1 and -7.6 kcal mol-1 and with the docking poses determined underneath the first α1-helix of the protein. Moreover, the molecular interaction determined by molecular dynamic (MD) simulation revealed that renieroside C1 (60) is more conveniently involved in key hydrophobic interactions with the best stability, least deviation, least ΔG (-6.9 kcal mol-1) and an RMSD value of 3.6 Å. Thus, the structural insights assure better binding affinity and favorable molecular interaction of renieroside C1 (60) towards the hACE2 protein, which plays a crucial role in the biology and pathogenesis of SARS-CoV-2.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Computational Evidences of Phytochemical Mediated Disruption of PLpro Driven Replication of SARS-CoV-2: A Therapeutic Approach against COVID-19.Curr Pharm Biotechnol. 2021;22(10):1350-1359. doi: 10.2174/1389201021999201110204116. Curr Pharm Biotechnol. 2021. PMID: 33176643
-
Active site-specific quantum tunneling of hACE2 receptor to assess its complexing poses with selective bioactive compounds in co-suppressing SARS-CoV-2 influx and subsequent cardiac injury.J Adv Vet Anim Res. 2021 Sep 29;8(4):540-556. doi: 10.5455/javar.2021.h544. eCollection 2021 Dec. J Adv Vet Anim Res. 2021. PMID: 35106293 Free PMC article.
-
Identification of bioactive molecule from Withania somnifera (Ashwagandha) as SARS-CoV-2 main protease inhibitor.J Biomol Struct Dyn. 2021 Sep;39(15):5668-5681. doi: 10.1080/07391102.2020.1790425. Epub 2020 Jul 8. J Biomol Struct Dyn. 2021. PMID: 32643552 Free PMC article.
-
Structure-based virtual screening and molecular dynamics simulation of SARS-CoV-2 Guanine-N7 methyltransferase (nsp14) for identifying antiviral inhibitors against COVID-19.J Biomol Struct Dyn. 2021 Aug;39(13):4582-4593. doi: 10.1080/07391102.2020.1778535. Epub 2020 Jun 22. J Biomol Struct Dyn. 2021. PMID: 32567979 Free PMC article.
-
Marine-Derived Bioactive Metabolites as a Potential Therapeutic Intervention in Managing Viral Diseases: Insights from the SARS-CoV-2 In Silico and Pre-Clinical Studies.Pharmaceuticals (Basel). 2024 Mar 1;17(3):328. doi: 10.3390/ph17030328. Pharmaceuticals (Basel). 2024. PMID: 38543114 Free PMC article. Review.
Cited by
-
Chemical Constituents and Anticancer Activities of Marine-Derived Fungus Trichoderma lixii.Molecules. 2024 Apr 29;29(9):2048. doi: 10.3390/molecules29092048. Molecules. 2024. PMID: 38731537 Free PMC article.
-
Evaluation of antiviral activity of Carica papaya leaves against SARS-CoV-2 assisted by metabolomic profiling.RSC Adv. 2022 Nov 16;12(51):32844-32852. doi: 10.1039/d2ra04600h. eCollection 2022 Nov 15. RSC Adv. 2022. PMID: 36425179 Free PMC article.
-
New Halogenated Compounds from Halimeda macroloba Seaweed with Potential Inhibitory Activity against Malaria.Molecules. 2022 Aug 31;27(17):5617. doi: 10.3390/molecules27175617. Molecules. 2022. PMID: 36080381 Free PMC article.
-
Wound Restorative Power of Halimeda macroloba/ Mesenchymal Stem Cells in Immunocompromised Rats via Downregulating Inflammatory/Immune Cross Talk.Mar Drugs. 2023 May 30;21(6):336. doi: 10.3390/md21060336. Mar Drugs. 2023. PMID: 37367661 Free PMC article.
-
Aurasperone A Inhibits SARS CoV-2 In Vitro: An Integrated In Vitro and In Silico Study.Mar Drugs. 2022 Feb 28;20(3):179. doi: 10.3390/md20030179. Mar Drugs. 2022. PMID: 35323478 Free PMC article.
References
-
- Wu Z. J. Ouyang M. A. Su R. K. Guo Y. X. Chin. J. Chem. 2008;26:759–764. doi: 10.1002/cjoc.200890142. - DOI
-
- Costantino V. de Rosa C. Fattorusso E. Imperatore C. Mangoni A. Irace C. Maffettone C. Capasso D. Malorni L. Palumbo R. Eur. J. Org. Chem. 2007;2007:5277–5283. doi: 10.1002/ejoc.200700390. - DOI
Publication types
LinkOut - more resources
Full Text Sources
Miscellaneous

