Uncovering N4-Acetylcytidine-Related mRNA Modification Pattern and Landscape of Stemness and Immunity in Hepatocellular Carcinoma
- PMID: 35493106
- PMCID: PMC9046676
- DOI: 10.3389/fcell.2022.861000
Uncovering N4-Acetylcytidine-Related mRNA Modification Pattern and Landscape of Stemness and Immunity in Hepatocellular Carcinoma
Abstract
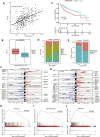
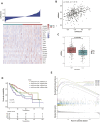
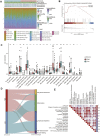
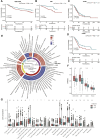
N4-acetylcytidine (ac4C) is an ancient and conserved RNA modification. Previously, ac4C mRNA modification has been reported promoting proliferation and metastasis of tumor cells. However, it remains unclear whether and how ac4C-related mRNA modification patterns influencing the prognosis of hepatocellular carcinoma (HCC) patients. Hereby, we constructed an ac4Cscore model and classified patients into two groups and investigated the potential intrinsic and extrinsic characteristics of tumor. The ac4Cscore model, including COL15A1, G6PD and TP53I3, represented ac4C-related mRNA modification patterns in HCC. According to ac4Cscore, patients were stratified to high and low groups with distinct prognosis. Patients subject to high group was related to advanced tumor stage, higher TP53 mutation rate, higher tumor stemness, more activated pathways in DNA-repair system, lower stromal score, higher immune score and higher infiltrating of T cells regulatory. While patients attributed to low group were correlated with abundance of T cells CD4 memory, less aggressive immune subtype and durable therapy benefit. We also found ac4Cscore as a novel marker to predict patients' prognosis with anti-PD1 immunotherapy and/or mTOR inhibitor treatment. Our study for the first time showed the association between ac4C-related mRNA modification patterns and tumor intrinsic and extrinsic characteristics, thus influencing the prognosis of patients.
Keywords: ac4C; hepatocellular carcinoma; mRNA modification; prognosis predictor; tumor microenvironment; tumor stemness.
Copyright © 2022 Liu, Zhang, Qiu, Zhang, Meng, Huang, Chen, Zhang and Han.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

