Immune Gene Signatures and Immunotypes in Immune Microenvironment Are Associated With Glioma Prognose
- PMID: 35493457
- PMCID: PMC9046586
- DOI: 10.3389/fimmu.2022.823910
Immune Gene Signatures and Immunotypes in Immune Microenvironment Are Associated With Glioma Prognose
Abstract
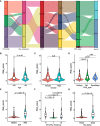
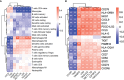
Glioma is the most common primary malignant brain tumor in adults with very poor prognosis. The limited new therapeutic strategies for glioma patients can be partially attributed to the complex tumor microenvironment. However, knowledge about the glioma immune microenvironment and the associated regulatory mechanisms is still lacking. In this study, we found that, different immune subtypes have a significant impact on patient survival. Glioma patients with a high immune response subtype had a shorter survival compared with patients with a low immune response subtype. Moreover, the number of B cell, T cell, NK cell, and in particular, the macrophage in the immune microenvironment of patients with a high immune response subtype were significantly enhanced. In addition, 132 genes were found to be related to glioma immunity. The functional analysis and verification of seven core genes showed that their expression levels were significantly correlated with the prognosis of glioma patients, and the results were consistent at tissue levels. These findings indicated that the glioma immune microenvironment was significantly correlated with the prognosis of glioma patients and multiple genes were involved in regulating the progression of glioma. The identified genes could be used to stratify glioma patients based on immune subgroup analysis, which may guide their clinical treatment regimen.
Keywords: glioma; immune microenvironment; immunotherapy; immunotype; macrophages.
Copyright © 2022 Wang, Cao, Zhai, Deng, Chao, Hu, Mou, Guo, Zhao, Li, Jiao, Xue, Han, Zhang and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







References
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

