Molecular Approaches for the Validation of the Baboon as a Nonhuman Primate Model for the Study of Zika Virus Infection
- PMID: 35493734
- PMCID: PMC9046911
- DOI: 10.3389/fcimb.2022.880860
Molecular Approaches for the Validation of the Baboon as a Nonhuman Primate Model for the Study of Zika Virus Infection
Abstract
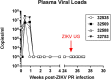
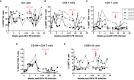
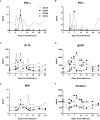
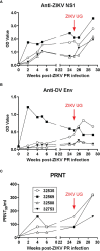
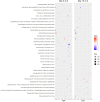
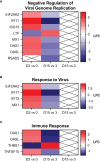
Nonhuman primates (NHP) are particularly important for modeling infections with viruses that do not naturally replicate in rodent cells. Zika virus (ZIKV) has been responsible for sporadic epidemics, but in 2015 a disseminated outbreak of ZIKV resulted in the World Health Organization declaring it a global health emergency. Since the advent of this last epidemic, several NHP species, including the baboon, have been utilized for modeling and understanding the complications of ZIKV infection in humans; several health issues related to the outcome of infection have not been resolved yet and require further investigation. This study was designed to validate, in baboons, the molecular signatures that have previously been identified in ZIKV-infected humans and macaque models. We performed a comprehensive molecular analysis of baboons during acute ZIKV infection, including flow cytometry, cytokine, immunological, and transcriptomic analyses. We show here that, similar to most human cases, ZIKV infection of male baboons tends to be subclinical, but is associated with a rapid and transient antiviral interferon-based response signature that induces a detectable humoral and cell-mediated immune response. This immunity against the virus protects animals from challenge with a divergent ZIKV strain, as evidenced by undetectable viremia but clear anamnestic responses. These results provide additional support for the use of baboons as an alternative animal model to macaques and validate omic techniques that could help identify the molecular basis of complications associated with ZIKV infections in humans.
Keywords: arboviral diseases; cytokines; immunology and infectious diseases; nonhuman primate (NHP); transcriptomic.
Copyright © 2022 Mask, Hodara, Callery, Parodi, Obregon-Perko, Yagi, Glenn, Frost, Clemmons, Patterson, Cox and Giavedoni.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

