Piper DNA virus 1 and 2 are endogenous pararetroviruses integrated into chromosomes of black pepper (Piper nigrum L)
- PMID: 35493754
- PMCID: PMC9005556
- DOI: 10.1007/s13337-021-00752-w
Piper DNA virus 1 and 2 are endogenous pararetroviruses integrated into chromosomes of black pepper (Piper nigrum L)
Abstract
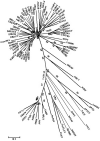
A previous study named 7178 and 892 bp contigs obtained through high-throughput sequencing (HTS) of black pepper as Piper DNA virus 1 (PDV-1) and PDV-2 respectively. In the present study, HTS results were confirmed through polymerase chain reaction and Sanger sequencing. The sequenced region of both PDV-1 and PDV-2 contained partial genomes with motifs characteristic of pararetroviruses. BLAST analysis of PDV-1 and PDV-2 against the whole genome sequence of the black pepper showed integration of the PDV-1 at 22 loci in chromosome number 14, and PDV-2 at two loci in chromosome number 12 of black pepper. The integration was confirmed through amplification and sequencing of the junction regions. The present study suggests that both PDV-1 and PDV-2 occur as endogenous viruses in black pepper. Further studies are needed to determine whether these endogenous viruses occur in episomal forms, their complete genome sequence and whether they are activable under abiotic stress conditions.
Supplementary information: The online version contains supplementary material available at 10.1007/s13337-021-00752-w.
Keywords: Caulimoviridae; Integrated virus; PCR; Sequencing; Tungrovirus.
© The Author(s), under exclusive licence to Indian Virological Society 2022.
Conflict of interest statement
Conflict of interestAll authors declare that they have no conflict of interest.
Figures

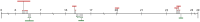
References
-
- Bhat AI, Siljo A, Jiby MV, Thankamani CK, Mathew PA. Polymerase chain reaction (PCR) based indexing for screening black pepper (Piper nigrum L.) plants against Piper yellow mottle virus. J Spices Aromatic Crops. 2009;18:28–32.
LinkOut - more resources
Full Text Sources
Research Materials

