Deep neural network modeling identifies biomarkers of response to immune-checkpoint therapy
- PMID: 35494249
- PMCID: PMC9044175
- DOI: 10.1016/j.isci.2022.104228
Deep neural network modeling identifies biomarkers of response to immune-checkpoint therapy
Abstract
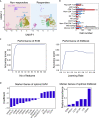
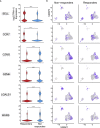
Immunotherapy has shown significant promise as a treatment for cancer, such as lung cancer and melanoma. However, only 10%-30% of the patients respond to treatment with immune checkpoint blockers (ICBs), underscoring the need for biomarkers to predict response to immunotherapy. Here, we developed DeepGeneX, a computational framework that uses advanced deep neural network modeling and feature elimination to reduce single-cell RNA-seq data on ∼26,000 genes to six of the most important genes (CCR7, SELL, GZMB, WARS, GZMH, and LGALS1), that accurately predict response to immunotherapy. We also discovered that the high LGALS1 and WARS-expressing macrophage population represent a biomarker for ICB therapy nonresponders, suggesting that these macrophages may be a target for improving ICB response. Taken together, DeepGeneX enables biomarker discovery and provides an understanding of the molecular basis for the model's predictions.
Keywords: Artificial intelligence; Cancer; Gene network; Immune response; Neural networks.
© 2022 The Author(s).
Conflict of interest statement
TSG and SV have filed for patent protection on the DeepGeneX approach. No other authors have any competing interests. TSG serves on the editorial board of iScience.
Figures




References
-
- Browaeys R., Saelens W., Saeys Y. NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods. 2020;17:159–162. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

