NCP-BiRW: A Hybrid Approach for Predicting Long Noncoding RNA-Disease Associations by Network Consistency Projection and Bi-Random Walk
- PMID: 35495166
- PMCID: PMC9043107
- DOI: 10.3389/fgene.2022.862272
NCP-BiRW: A Hybrid Approach for Predicting Long Noncoding RNA-Disease Associations by Network Consistency Projection and Bi-Random Walk
Abstract
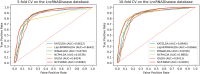
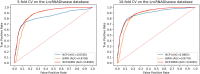
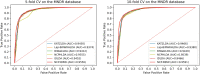
Long non-coding RNAs (lncRNAs) play significant roles in the disease process. Understanding the pathological mechanisms of lncRNAs during the course of various diseases will help clinicians prevent and treat diseases. With the emergence of high-throughput techniques, many biological experiments have been developed to study lncRNA-disease associations. Because experimental methods are costly, slow, and laborious, a growing number of computational models have emerged. Here, we present a new approach using network consistency projection and bi-random walk (NCP-BiRW) to infer hidden lncRNA-disease associations. First, integrated similarity networks for lncRNAs and diseases were constructed by merging similarity information. Subsequently, network consistency projection was applied to calculate space projection scores for lncRNAs and diseases, which were then introduced into a bi-random walk method for association prediction. To test model performance, we employed 5- and 10-fold cross-validation, with the area under the receiver operating characteristic curve as the evaluation indicator. The computational results showed that our method outperformed the other five advanced algorithms. In addition, the novel method was applied to another dataset in the Mammalian ncRNA-Disease Repository (MNDR) database and showed excellent performance. Finally, case studies were carried out on atherosclerosis and leukemia to confirm the effectiveness of our method in practice. In conclusion, we could infer lncRNA-disease associations using the NCP-BiRW model, which may benefit biomedical studies in the future.
Keywords: bi-random walk; integrated similarity; lncRNA-disease association prediction; network consistency projection; normalization.
Copyright © 2022 Liu, Yang, Zheng, Wang, Yan, Cao and Zhang.
Conflict of interest statement
The authors declare that the research was conducted without any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
Similar articles
-
Laplacian normalization and bi-random walks on heterogeneous networks for predicting lncRNA-disease associations.BMC Syst Biol. 2018 Dec 31;12(Suppl 9):122. doi: 10.1186/s12918-018-0660-0. BMC Syst Biol. 2018. PMID: 30598088 Free PMC article.
-
BRWMC: Predicting lncRNA-disease associations based on bi-random walk and matrix completion on disease and lncRNA networks.Comput Biol Chem. 2023 Apr;103:107833. doi: 10.1016/j.compbiolchem.2023.107833. Epub 2023 Feb 17. Comput Biol Chem. 2023. PMID: 36812824
-
A novel target convergence set based random walk with restart for prediction of potential LncRNA-disease associations.BMC Bioinformatics. 2019 Dec 3;20(1):626. doi: 10.1186/s12859-019-3216-4. BMC Bioinformatics. 2019. PMID: 31795943 Free PMC article.
-
RWSF-BLP: a novel lncRNA-disease association prediction model using random walk-based multi-similarity fusion and bidirectional label propagation.Mol Genet Genomics. 2021 May;296(3):473-483. doi: 10.1007/s00438-021-01764-3. Epub 2021 Feb 15. Mol Genet Genomics. 2021. PMID: 33590345 Review.
-
Long non-coding RNAs and complex diseases: from experimental results to computational models.Brief Bioinform. 2017 Jul 1;18(4):558-576. doi: 10.1093/bib/bbw060. Brief Bioinform. 2017. PMID: 27345524 Free PMC article. Review.
Cited by
-
Predicting lncRNA-Disease Associations Based on a Dual-Path Feature Extraction Network with Multiple Sources of Information Integration.ACS Omega. 2024 Jul 30;9(32):35100-35112. doi: 10.1021/acsomega.4c05365. eCollection 2024 Aug 13. ACS Omega. 2024. PMID: 39157140 Free PMC article.
References
LinkOut - more resources
Full Text Sources

