LncRNA NEAT1 Potentiates SREBP2 Activity to Promote Inflammatory Macrophage Activation and Limit Hantaan Virus Propagation
- PMID: 35495674
- PMCID: PMC9044491
- DOI: 10.3389/fmicb.2022.849020
LncRNA NEAT1 Potentiates SREBP2 Activity to Promote Inflammatory Macrophage Activation and Limit Hantaan Virus Propagation
Abstract
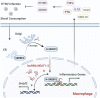
As the global prototypical zoonotic hantavirus, Hantaan virus (HTNV) is prevalent in Asia and is the leading causative agent of severe hemorrhagic fever with renal syndrome (HFRS), which has profound morbidity and mortality. Macrophages are crucial components of the host innate immune system and serve as the first line of defense against HTNV infection. Previous studies indicated that the viral replication efficiency in macrophages determines hantavirus pathogenicity, but it remains unknown which factor manipulates the macrophage activation pattern and the virus-host interaction process. Here, we performed the transcriptomic analysis of HTNV-infected mouse bone marrow-derived macrophages and identified the long noncoding RNA (lncRNA) nuclear enriched abundant transcript 1 (NEAT1), especially the isoform NEAT1-2, as one of the lncRNAs that is differentially expressed at the early phase. Based on coculture experiments, we revealed that silencing NEAT1-2 hinders inflammatory macrophage activation and facilitates HTNV propagation, while enhancing NEAT1-2 transcription effectively restrains viral replication. Furthermore, sterol response element binding factor-2 (SREBP2), which controls the cholesterol metabolism process, was found to stimulate macrophages by promoting the production of multiple inflammatory cytokines upon HTNV infection. NEAT1-2 could potentiate SREBP2 activity by upregulating Srebf1 expression and interacting with SREBP2, thus stimulating inflammatory macrophages and limiting HTNV propagation. More importantly, we demonstrated that the NEAT1-2 expression level in patient monocytes was negatively correlated with viral load and HFRS disease progression. Our results identified a function and mechanism of action for the lncRNA NEAT1 in heightening SREBP2-mediated macrophage activation to restrain hantaviral propagation and revealed the association of NEAT1 with HFRS severity.
Keywords: HFRS; Hantaan virus; NEAT1; SREBP2; hantavirus; inflammatory macrophage; lncRNA.
Copyright © 2022 Yang, Li, Ma, Ye, Si, Zheng, Liu, Cheng, Zhang, Zhang, Zhang, Lei, Shen, Zhang and Ma.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Chinese National Bureau of Statistics (2020). The epidemiology feature of HFRS in China. Available at: http://www.stats.gov.cn/ (Accessed October 10, 2020).
LinkOut - more resources
Full Text Sources
Research Materials

