Real-time detection of mRNA splicing variants with specifically designed reverse-transcription loop-mediated isothermal amplification
- PMID: 35495989
- PMCID: PMC9049701
- DOI: 10.1039/d0ra00591f
Real-time detection of mRNA splicing variants with specifically designed reverse-transcription loop-mediated isothermal amplification
Abstract
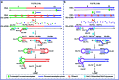
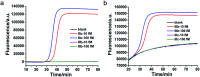
Alternative splicing is a ubiquitous and crucial process in cellular processes and has a specific linkage with diseases. To date, developing cost-effective methods with high sensitivity and specificity for detection of splicing variants has been needed. Herein, we report a novel splicing variant assay based on specifically designed reverse-transcription loop-mediated isothermal amplification. After reverse transcribing the splicing variant into cDNA, four DNA primers are specifically designed to recognize six distinct regions. The four DNA primers can hybridize with corresponding sequences for extension and strand displacement DNA synthesis to form stem-loop DNA and then LAMP amplification is started. The proposed method can detect as low as 100 aM splicing variants in real-time fashion with high specificity, showing great potential in biological function and clinical studies.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts to declare.
Figures


Similar articles
-
Specific recognition and sensitive quantification of mRNA splice variants via one-pot ligation-dependent loop-mediated isothermal amplification.Analyst. 2023 Nov 6;148(22):5605-5611. doi: 10.1039/d3an01382k. Analyst. 2023. PMID: 37818948
-
Loop-mediated isothermal amplification of DNA.Nucleic Acids Res. 2000 Jun 15;28(12):E63. doi: 10.1093/nar/28.12.e63. Nucleic Acids Res. 2000. PMID: 10871386 Free PMC article.
-
Immunocapture-Reverse Transcriptase Loop-Mediated Isothermal Amplification Assay for Detection of Plant RNA Viruses.Methods Mol Biol. 2022;2400:245-252. doi: 10.1007/978-1-0716-1835-6_23. Methods Mol Biol. 2022. PMID: 34905207
-
Development and evaluation of an in-house single step loop-mediated isothermal amplification (SS-LAMP) assay for the detection of Mycobacterium tuberculosis complex in sputum samples from Moroccan patients.BMC Infect Dis. 2016 Sep 27;16(1):517. doi: 10.1186/s12879-016-1864-9. BMC Infect Dis. 2016. PMID: 27677540 Free PMC article.
-
Loop-mediated isothermal amplification (LAMP): a versatile technique for detection of micro-organisms.J Appl Microbiol. 2018 Mar;124(3):626-643. doi: 10.1111/jam.13647. Epub 2018 Feb 12. J Appl Microbiol. 2018. PMID: 29165905 Free PMC article. Review.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources

