Protocol to assess human glioma propagating cell migration on linear micropatterns mimicking brain invasion tracks
- PMID: 35496779
- PMCID: PMC9043773
- DOI: 10.1016/j.xpro.2022.101331
Protocol to assess human glioma propagating cell migration on linear micropatterns mimicking brain invasion tracks
Abstract
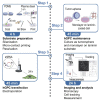
Glioblastoma (GBM) cells invade the brain by following linear structures like blood vessel walls and white matter tracts by using specific motility modes. In this protocol, we describe two micropatterning techniques allowing recapitulation of these linear tracks in vitro: micro-contact printing and deep UV photolithography. We also detail how to maintain, transfect, and prepare human glioma propagating cells (hGPCs) for migration assays on linear tracks, followed by image acquisition and analysis, to measure key parameters of their motility. For complete details on the use and execution of this protocol, please refer to Monzo et al. (2016) and Monzo et al. (2021a).
Keywords: Biophysics; Biotechnology and bioengineering; Cancer; Cell Biology; Cell culture; Cell-based Assays; Microscopy.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures





Similar articles
-
Protocol using ex vivo mouse brain slice culture mimicking in vivo conditions to study tumor growth and cell motility of glioblastoma cells.STAR Protoc. 2024 Dec 20;5(4):103401. doi: 10.1016/j.xpro.2024.103401. Epub 2024 Oct 19. STAR Protoc. 2024. PMID: 39425931 Free PMC article.
-
Mechanical confinement triggers glioma linear migration dependent on formin FHOD3.Mol Biol Cell. 2016 Apr 15;27(8):1246-61. doi: 10.1091/mbc.E15-08-0565. Epub 2016 Feb 24. Mol Biol Cell. 2016. PMID: 26912794 Free PMC article.
-
Glioma Cell Migration Dynamics in Brain Tissue Assessed by Multimodal Optical Imaging.Biophys J. 2019 Oct 1;117(7):1179-1188. doi: 10.1016/j.bpj.2019.08.010. Epub 2019 Aug 15. Biophys J. 2019. PMID: 31474305 Free PMC article.
-
Mechanisms regulating glioma invasion.Cancer Lett. 2015 Jun 28;362(1):1-7. doi: 10.1016/j.canlet.2015.03.015. Epub 2015 Mar 18. Cancer Lett. 2015. PMID: 25796440 Free PMC article. Review.
-
Integrative analysis of cell adhesion molecules in glioblastoma identified prostaglandin F2 receptor inhibitor (PTGFRN) as an essential gene.BMC Cancer. 2022 Jun 11;22(1):642. doi: 10.1186/s12885-022-09682-2. BMC Cancer. 2022. PMID: 35690717 Free PMC article. Review.
Cited by
-
Engineering spatially-confined conduits to tune nerve self-organization and allodynic responses via YAP-mediated mechanotransduction.Nat Commun. 2025 Jan 2;16(1):66. doi: 10.1038/s41467-024-55118-9. Nat Commun. 2025. PMID: 39746959 Free PMC article.
-
Development of label-free light-controlled gene expression technologies using mid-IR and terahertz light.Front Bioeng Biotechnol. 2024 Oct 11;12:1324757. doi: 10.3389/fbioe.2024.1324757. eCollection 2024. Front Bioeng Biotechnol. 2024. PMID: 39465004 Free PMC article. Review.
References
-
- Azioune A., Carpi N., Tseng Q., Thery M., Piel M. Protein micropatterns: a direct printing protocol using deep UVs. Methods Cell Biol. 2010;97:133–146. - PubMed
-
- Azioune A., Storch M., Bornens M., Thery M., Piel M. Simple and rapid process for single cell micro-patterning. Lab Chip. 2009;9:1640–1642. - PubMed
-
- Bellail A.C., Hunter S.B., Brat D.J., Tan C., Van Meir E.G. Microregional extracellular matrix heterogeneity in brain modulates glioma cell invasion. Int. J. Biochem. Cell Biol. 2004;36:1046–1069. - PubMed
-
- Bellon G., Caulet T., Cam Y., Pluot M., Poulin G., Pytlinska M., Bernard M.H. Immunohistochemical localisation of macromolecules of the basement membrane and extracellular matrix of human gliomas and meningiomas. Acta Neuropathol. 1985;66:245–252. - PubMed
-
- Charles N.A., Holland E.C., Gilbertson R., Glass R., Kettenmann H. The brain tumor microenvironment. Glia. 2012;60:502–514. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Medical

