Inflammatory bowel disease biomarkers of human gut microbiota selected via different feature selection methods
- PMID: 35497193
- PMCID: PMC9048649
- DOI: 10.7717/peerj.13205
Inflammatory bowel disease biomarkers of human gut microbiota selected via different feature selection methods
Abstract
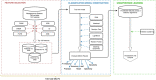
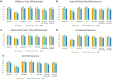
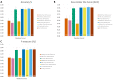
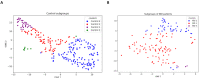
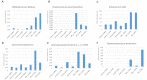
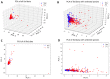
The tremendous boost in next generation sequencing and in the "omics" technologies makes it possible to characterize the human gut microbiome-the collective genomes of the microbial community that reside in our gastrointestinal tract. Although some of these microorganisms are considered to be essential regulators of our immune system, the alteration of the complexity and eubiotic state of microbiota might promote autoimmune and inflammatory disorders such as diabetes, rheumatoid arthritis, Inflammatory bowel diseases (IBD), obesity, and carcinogenesis. IBD, comprising Crohn's disease and ulcerative colitis, is a gut-related, multifactorial disease with an unknown etiology. IBD presents defects in the detection and control of the gut microbiota, associated with unbalanced immune reactions, genetic mutations that confer susceptibility to the disease, and complex environmental conditions such as westernized lifestyle. Although some existing studies attempt to unveil the composition and functional capacity of the gut microbiome in relation to IBD diseases, a comprehensive picture of the gut microbiome in IBD patients is far from being complete. Due to the complexity of metagenomic studies, the applications of the state-of-the-art machine learning techniques became popular to address a wide range of questions in the field of metagenomic data analysis. In this regard, using IBD associated metagenomics dataset, this study utilizes both supervised and unsupervised machine learning algorithms, (i) to generate a classification model that aids IBD diagnosis, (ii) to discover IBD-associated biomarkers, (iii) to discover subgroups of IBD patients using k-means and hierarchical clustering approaches. To deal with the high dimensionality of features, we applied robust feature selection algorithms such as Conditional Mutual Information Maximization (CMIM), Fast Correlation Based Filter (FCBF), min redundancy max relevance (mRMR), Select K Best (SKB), Information Gain (IG) and Extreme Gradient Boosting (XGBoost). In our experiments with 100-fold Monte Carlo cross-validation (MCCV), XGBoost, IG, and SKB methods showed a considerable effect in terms of minimizing the microbiota used for the diagnosis of IBD and thus reducing the cost and time. We observed that compared to Decision Tree, Support Vector Machine, Logitboost, Adaboost, and stacking ensemble classifiers, our Random Forest classifier resulted in better performance measures for the classification of IBD. Our findings revealed potential microbiome-mediated mechanisms of IBD and these findings might be useful for the development of microbiome-based diagnostics.
Keywords: Biomarker discovery; Classification; Feature selection; Human gut microbiome; Metagenomics.
©2022 Bakir-Gungor et al.
Conflict of interest statement
Burcu Bakir-Gungor is an Academic Editor for PeerJ.
Figures




References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

