An integrated toolbox to profile macrophage immunometabolism
- PMID: 35497494
- PMCID: PMC9046227
- DOI: 10.1016/j.crmeth.2022.100192
An integrated toolbox to profile macrophage immunometabolism
Abstract
Macrophages are dynamic immune cells that can adopt several activation states. Fundamental to these functional activation states is the regulation of cellular metabolic processes. Especially in mice, metabolic alterations underlying pro-inflammatory or homeostatic phenotypes have been assessed using various techniques. However, researchers new to the field may encounter ambiguity in choosing which combination of techniques is best suited to profile immunometabolism. To address this need, we have developed a toolbox to assess cellular metabolism in a semi-high-throughput 96-well-plate-based format. Application of the toolbox to activated mouse and human macrophages enables fast metabolic pre-screening and robust measurement of extracellular fluxes, mitochondrial mass and membrane potential, and glucose and lipid uptake. Moreover, we propose an application of SCENITH technology for ex vivo metabolic profiling. We validate established activation-induced metabolic rewiring in mouse macrophages and report new insights into human macrophage metabolism. By thoroughly discussing each technique, we hope to guide readers with practical workflows for investigating immunometabolism.
Keywords: immunometabolism; macrophages; metabolism; semi-high throughput screening; toolbox.
© 2022 The Author(s).
Conflict of interest statement
The authors declare no competing interests. There are restrictions to the commercial use of SCENITH due to a pending patent application (PCT/EP2020/060,486).
Figures




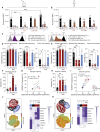
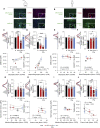
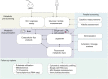
References
-
- Arguello R.J., Combes A.J., Char R., Gigan J.P., Baaziz A.I., Bousiquot E., Camosseto V., Samad B., Tsui J., Yan P., et al. SCENITH: a flow cytometry-based method to functionally profile energy metabolism with single-cell resolution. Cell Metab. 2020;32:1063–1075.e67. doi: 10.1016/j.cmet.2020.11.007. - DOI - PMC - PubMed
-
- Baardman J., Verberk S.G.S., Prange K.H.M., van Weeghel M., van der Velden S., Ryan D.G., Wust R.C.I., Neele A.E., Speijer D., Denis S.W., et al. A defective pentose phosphate pathway reduces inflammatory macrophage responses during hypercholesterolemia. Cell Rep. 2018;25:2044–2052.e45. doi: 10.1016/j.celrep.2018.10.092. - DOI - PubMed
-
- Bailey J.D., Diotallevi M., Nicol T., McNeill E., Shaw A., Chuaiphichai S., Hale A., Starr A., Nandi M., Stylianou E., et al. Nitric oxide modulates metabolic remodeling in inflammatory macrophages through TCA cycle regulation and itaconate accumulation. Cell Rep. 2019;28:218–230.e17. doi: 10.1016/j.celrep.2019.06.018. - DOI - PMC - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources

