Characterization of Genetic Variants of Uncertain Significance for the ALPL Gene in Patients With Adult Hypophosphatasia
- PMID: 35498405
- PMCID: PMC9047899
- DOI: 10.3389/fendo.2022.863940
Characterization of Genetic Variants of Uncertain Significance for the ALPL Gene in Patients With Adult Hypophosphatasia
Abstract
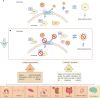
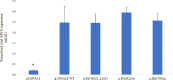
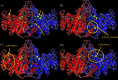
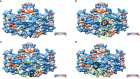
Hypophosphatasia (HPP) a rare disease caused by mutations in the ALPL gene encoding for the tissue-nonspecific alkaline phosphatase protein (TNSALP), has been identified as a potentially under-diagnosed condition worldwide which may have higher prevalence than currently established. This is largely due to the overlapping of its symptomatology with that of other more frequent pathologies. Although HPP is usually associated with deficient bone mineralization, the high genetic variability of ALPL results in high clinical heterogeneity, which makes it difficult to establish a specific HPP symptomatology. In the present study, three variants of ALPL gene with uncertain significance and no previously described (p.Del Glu23_Lys24, p.Pro292Leu and p.His379Asn) were identified in heterozygosis in patients diagnosed with HPP. These variants were characterized at phenotypic, functional and structural levels. All genetic variants showed significantly lower in vitro ALP activity than the wild-type (WT) genotype (p-value <0.001). Structurally, p.His379Asn variant resulted in the loss of two Zn2+ binding sites in the protein dimer which may greatly affect ALP activity. In summary, we identified three novel ALPL gene mutations associated with adult HPP. The correct identification and characterization of new variants and the subsequent study of their phenotype will allow the establishment of genotype-phenotype relationships that facilitate the management of the disease as well as making it possible to individualize treatment for each specific patient. This would allow the therapeutic approach to HPP to be personalized according to the unique genetic characteristics and clinical manifestations of each patient.
Keywords: alkaline phosphatase; bone; enzymatic activity; genetic variant; hypophosphatasia; mineralization; pyridoxal 5´ phosphate.
Copyright © 2022 Sanabria-de la Torre, Martínez-Heredia, González-Salvatierra, Andújar-Vera, Iglesias-Baena, Villa-Suárez, Contreras-Bolívar, Corbacho-Soto, Martínez-Navajas, Real, García-Fontana, Muñoz-Torres and García-Fontana.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- García-Fontana C, Villa-Suárez JM, Andújar-Vera F, González-Salvatierra S, Martínez-Navajas G, Real PJ, et al. . Epidemiological, Clinical and Genetic Study of Hypophosphatasia in A Spanish Population: Identification of Two Novel Mutations in The Alpl Gene. Sci Rep (2019) 9:1–11. doi: 10.1038/s41598-019-46004-2 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
LinkOut - more resources
Full Text Sources
Miscellaneous

