The Use and Limitations of Exome Capture to Detect Novel Variation in the Hexaploid Wheat Genome
- PMID: 35498663
- PMCID: PMC9039655
- DOI: 10.3389/fpls.2022.841855
The Use and Limitations of Exome Capture to Detect Novel Variation in the Hexaploid Wheat Genome
Abstract
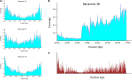
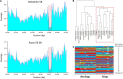
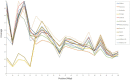
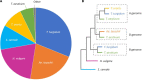
The bread wheat (Triticum aestivum) pangenome is a patchwork of variable regions, including translocations and introgressions from progenitors and wild relatives. Although a large number of these have been documented, it is likely that many more remain unknown. To map these variable regions and make them more traceable in breeding programs, wheat accessions need to be genotyped or sequenced. The wheat genome is large and complex and consequently, sequencing efforts are often targeted through exome capture. In this study, we employed exome capture prior to sequencing 12 wheat varieties; 10 elite T. aestivum cultivars and two T. aestivum landrace accessions. Sequence coverage across chromosomes was greater toward distal regions of chromosome arms and lower in centromeric regions, reflecting the capture probe distribution which itself is determined by the known telomere to centromere gene gradient. Superimposed on this general pattern, numerous drops in sequence coverage were observed. Several of these corresponded with reported introgressions. Other drops in coverage could not be readily explained and may point to introgressions that have not, to date, been documented.
Keywords: Triticum aestivum; exome capture; exome capture sequencing; introgression; sequence variation; wheat.
Copyright © 2022 Burridge, Winfield, Wilkinson, Przewieslik-Allen, Edwards and Barker.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


References
-
- Allen A. M., Winfield M. O., Burridge A. J., Downie R. C., Benbow H. R., Barker G. L. A. (2016). Characterization of a Wheat Breeders’ Array suitable for high-throughput SNP genotyping of global accessions of hexaploid wheat (Triticum aestivum). Plant Biotechnol. J. 15 390–401. 10.1111/pbi.12635 - DOI - PMC - PubMed
-
- Arbor Biosciences (2021). myBaits® - Hybridization Capture for Targeted NGS – Manual v.5.01 - Long Insert Protocol. Ann Arbor, MI: Arbor Biosciences.
-
- Arraiano L. S., Chartrain L., Bossolini E., Slatter H. N., Keller B., Brown J. K. M. (2007). A gene in European wheat cultivars for resistance to an African isolate of Mycosphaerella graminicola. Plant Pathol. 56 73–78.
-
- Babraham Bioinformatics (2020). FastQC Available online at: https://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed June 9, 2020).
LinkOut - more resources
Full Text Sources

