Molecular Characterization Reveals the Involvement of Calcium Dependent Protein Kinases in Abiotic Stress Signaling and Development in Chickpea (Cicer arietinum)
- PMID: 35498712
- PMCID: PMC9039462
- DOI: 10.3389/fpls.2022.831265
Molecular Characterization Reveals the Involvement of Calcium Dependent Protein Kinases in Abiotic Stress Signaling and Development in Chickpea (Cicer arietinum)
Abstract
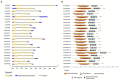
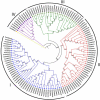
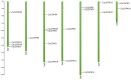
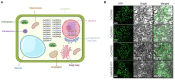
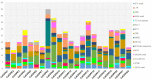
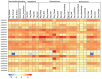
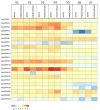
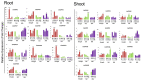
Calcium-dependent protein kinases (CDPKs) are a major group of calcium (Ca2+) sensors in plants. CDPKs play a dual function of "Ca2+ sensor and responder." These sensors decode the "Ca2+ signatures" generated in response to adverse growth conditions such as drought, salinity, and cold and developmental processes. However, knowledge of the CDPK family in the legume crop chickpea is missing. Here, we have identified a total of 22 CDPK genes in the chickpea genome. The phylogenetic analysis of the chickpea CDPK family with other plants revealed their evolutionary conservation. Protein homology modeling described the three-dimensional structure of chickpea CDPKs. Defined arrangements of α-helix, β-strands, and transmembrane-helix represent important structures like kinase domain, inhibitory junction domain, N and C-lobes of EF-hand motifs. Subcellular localization analysis revealed that CaCDPK proteins are localized mainly at the cytoplasm and in the nucleus. Most of the CaCDPK promoters had abiotic stress and development-related cis-regulatory elements, suggesting the functional role of CaCDPKs in abiotic stress and development-related signaling. RNA sequencing (RNA-seq) expression analysis indicated the role of the CaCDPK family in various developmental stages, including vegetative, reproductive development, senescence stages, and during seed stages of early embryogenesis, late embryogenesis, mid and late seed maturity. The real-time quantitative PCR (qRT-PCR) analysis revealed that several CaCDPK genes are specifically as well as commonly induced by drought, salt, and Abscisic acid (ABA). Overall, these findings indicate that the CDPK family is probably involved in abiotic stress responses and development in chickpeas. This study provides crucial information on the CDPK family that will be utilized in generating abiotic stress-tolerant and high-yielding chickpea varieties.
Keywords: CDPK; abiotic stress; chickpea; development; gene expression; signaling.
Copyright © 2022 Deepika, Poddar, Kumar and Singh.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Asai S., Ichikawa T., Nomura H., Kobayashi M., Kamiyoshihara Y., Mori H., et al. . (2013). The variable domain of a plant calcium-dependent protein kinase (CDPK) confers subcellular localization and substrate recognition for NADPH oxidase. J. Biol. Chem. 288, 14332–14340. 10.1074/jbc.M112.448910 - DOI - PMC - PubMed
-
- Asano T., Tanaka N., Yang G., Hayashi N., Komatsu S. (2005). Genome-wide identification of the rice calcium-dependent protein kinase. Plant Cell Physiol. 46, 356–366. - PubMed
LinkOut - more resources
Full Text Sources
Miscellaneous

